Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
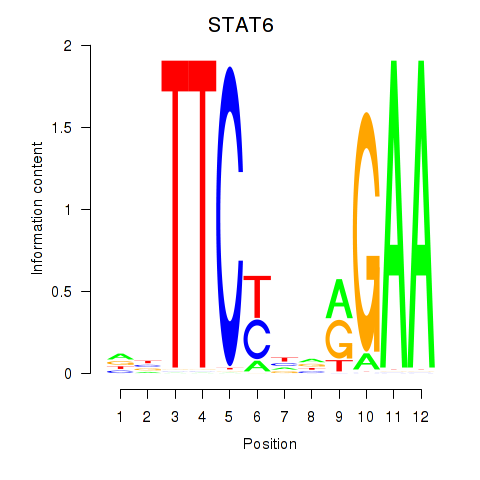
Results for STAT6
Z-value: 0.78
Transcription factors associated with STAT6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT6
|
ENSG00000166888.6 | signal transducer and activator of transcription 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT6 | hg19_v2_chr12_-_57505121_57505216 | 0.14 | 4.8e-01 | Click! |
Activity profile of STAT6 motif
Sorted Z-values of STAT6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_205391178 | 9.83 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chrX_+_135251783 | 3.14 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chrX_+_135252050 | 2.86 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chrX_+_135251835 | 2.85 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr2_+_113885138 | 2.68 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr10_+_102106829 | 2.53 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr2_-_31440377 | 2.18 |
ENST00000444918.2
ENST00000403897.3 |
CAPN14
|
calpain 14 |
| chr11_-_128457446 | 1.98 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr17_-_17740325 | 1.77 |
ENST00000338854.5
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr11_+_76745385 | 1.63 |
ENST00000533140.1
ENST00000354301.5 ENST00000528622.1 |
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr4_+_54966198 | 1.60 |
ENST00000326902.2
ENST00000503800.1 |
GSX2
|
GS homeobox 2 |
| chr9_+_36572851 | 1.58 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr22_+_44464923 | 1.53 |
ENST00000404989.1
|
PARVB
|
parvin, beta |
| chrX_-_106960285 | 1.48 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr8_+_49984894 | 1.45 |
ENST00000522267.1
ENST00000399653.4 ENST00000303202.8 |
C8orf22
|
chromosome 8 open reading frame 22 |
| chr1_-_160616804 | 1.41 |
ENST00000538290.1
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr2_-_190044480 | 1.36 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr11_+_65082289 | 1.29 |
ENST00000279249.2
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr12_+_9144626 | 1.23 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr14_+_55034599 | 1.19 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr19_+_38924316 | 1.13 |
ENST00000355481.4
ENST00000360985.3 ENST00000359596.3 |
RYR1
|
ryanodine receptor 1 (skeletal) |
| chr1_-_76398077 | 1.10 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr17_-_17740287 | 1.06 |
ENST00000355815.4
ENST00000261646.5 |
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr1_-_8086343 | 1.00 |
ENST00000474874.1
ENST00000469499.1 ENST00000377482.5 |
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr16_+_30075463 | 1.00 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_+_191792376 | 0.98 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr8_+_11666649 | 0.98 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr18_-_24445664 | 0.96 |
ENST00000578776.1
|
AQP4
|
aquaporin 4 |
| chr14_-_69446034 | 0.94 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr17_-_39507064 | 0.94 |
ENST00000007735.3
|
KRT33A
|
keratin 33A |
| chr1_+_150245099 | 0.88 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr1_-_241803679 | 0.86 |
ENST00000331838.5
|
OPN3
|
opsin 3 |
| chr6_-_27880174 | 0.85 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr2_+_182756915 | 0.85 |
ENST00000428267.2
|
SSFA2
|
sperm specific antigen 2 |
| chr16_+_30075595 | 0.84 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr11_-_67981046 | 0.84 |
ENST00000402789.1
ENST00000402185.2 ENST00000458496.1 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr6_-_30524951 | 0.83 |
ENST00000376621.3
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr1_+_150245177 | 0.82 |
ENST00000369098.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr1_-_41328018 | 0.82 |
ENST00000372638.2
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr1_-_209824643 | 0.77 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr16_+_30075783 | 0.76 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr12_+_6493319 | 0.75 |
ENST00000536876.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr3_-_98235962 | 0.75 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr19_+_2785458 | 0.73 |
ENST00000307741.6
ENST00000585338.1 |
THOP1
|
thimet oligopeptidase 1 |
| chr6_+_30525051 | 0.72 |
ENST00000376557.3
|
PRR3
|
proline rich 3 |
| chr11_+_112832133 | 0.72 |
ENST00000524665.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr2_+_173600514 | 0.70 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr4_-_120988229 | 0.69 |
ENST00000296509.6
|
MAD2L1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr2_+_173600565 | 0.69 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr3_+_140981456 | 0.69 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr3_-_10149907 | 0.68 |
ENST00000450660.2
ENST00000524279.1 |
FANCD2OS
|
FANCD2 opposite strand |
| chr3_+_158787041 | 0.67 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr2_+_219745020 | 0.66 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr6_+_38690622 | 0.66 |
ENST00000327475.6
|
DNAH8
|
dynein, axonemal, heavy chain 8 |
| chr3_+_189507523 | 0.65 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr4_+_72897521 | 0.65 |
ENST00000308744.6
ENST00000344413.5 |
NPFFR2
|
neuropeptide FF receptor 2 |
| chr2_+_68961934 | 0.64 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr6_-_117150198 | 0.64 |
ENST00000310357.3
ENST00000368549.3 ENST00000530250.1 |
GPRC6A
|
G protein-coupled receptor, family C, group 6, member A |
| chr3_-_73673991 | 0.64 |
ENST00000308537.4
ENST00000263666.4 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr20_+_58251716 | 0.63 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr2_+_68961905 | 0.62 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr18_-_24445729 | 0.58 |
ENST00000383168.4
|
AQP4
|
aquaporin 4 |
| chr9_-_131486367 | 0.58 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr1_+_214163033 | 0.57 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr2_-_204400113 | 0.57 |
ENST00000319170.5
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr11_-_14379997 | 0.56 |
ENST00000526063.1
ENST00000532814.1 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr1_+_32608566 | 0.56 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr9_-_14910990 | 0.55 |
ENST00000380881.4
ENST00000422223.2 |
FREM1
|
FRAS1 related extracellular matrix 1 |
| chr3_+_158991025 | 0.54 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr8_-_42623747 | 0.53 |
ENST00000534622.1
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr3_+_189507432 | 0.52 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr2_+_202316392 | 0.51 |
ENST00000194530.3
ENST00000392249.2 |
STRADB
|
STE20-related kinase adaptor beta |
| chr2_-_134326009 | 0.51 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr12_-_71031220 | 0.51 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr5_+_74011328 | 0.51 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr12_-_54779511 | 0.50 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr8_-_42623924 | 0.49 |
ENST00000276410.2
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr6_-_41254403 | 0.49 |
ENST00000589614.1
ENST00000334475.6 ENST00000591620.1 ENST00000244709.4 |
TREM1
|
triggering receptor expressed on myeloid cells 1 |
| chr19_-_54604083 | 0.48 |
ENST00000391761.1
ENST00000356532.3 ENST00000359649.4 ENST00000358375.4 ENST00000391760.1 ENST00000351806.4 |
OSCAR
|
osteoclast associated, immunoglobulin-like receptor |
| chr12_-_71031185 | 0.48 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr16_-_29757272 | 0.48 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr6_-_41168920 | 0.47 |
ENST00000483722.1
|
TREML2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr11_-_60719213 | 0.47 |
ENST00000227880.3
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chrX_-_138914394 | 0.47 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr7_+_129015484 | 0.46 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr15_+_91643442 | 0.45 |
ENST00000394232.1
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr5_+_126984710 | 0.45 |
ENST00000379445.3
|
CTXN3
|
cortexin 3 |
| chr15_+_89402148 | 0.45 |
ENST00000560601.1
|
ACAN
|
aggrecan |
| chr10_-_24770632 | 0.45 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr17_+_7384721 | 0.45 |
ENST00000412468.2
|
SLC35G6
|
solute carrier family 35, member G6 |
| chr2_+_182756615 | 0.44 |
ENST00000431877.2
ENST00000320370.7 |
SSFA2
|
sperm specific antigen 2 |
| chr1_-_205649580 | 0.42 |
ENST00000367145.3
|
SLC45A3
|
solute carrier family 45, member 3 |
| chr1_-_68299130 | 0.42 |
ENST00000370982.3
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12 |
| chr3_+_35682913 | 0.41 |
ENST00000449196.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr2_+_131769256 | 0.40 |
ENST00000355771.3
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr1_+_26759295 | 0.39 |
ENST00000430232.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr1_-_179834311 | 0.39 |
ENST00000553856.1
|
IFRG15
|
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr8_+_9953214 | 0.38 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr22_+_38201114 | 0.37 |
ENST00000340857.2
|
H1F0
|
H1 histone family, member 0 |
| chr12_-_82152444 | 0.37 |
ENST00000549325.1
ENST00000550584.2 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr16_-_67753206 | 0.36 |
ENST00000268797.7
|
GFOD2
|
glucose-fructose oxidoreductase domain containing 2 |
| chr20_+_42574317 | 0.36 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr14_-_37642016 | 0.35 |
ENST00000331299.5
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr2_+_197504278 | 0.35 |
ENST00000272831.7
ENST00000389175.4 ENST00000472405.2 ENST00000423093.2 |
CCDC150
|
coiled-coil domain containing 150 |
| chr8_+_9953061 | 0.33 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr12_-_53097247 | 0.33 |
ENST00000341809.3
ENST00000537195.1 |
KRT77
|
keratin 77 |
| chr11_+_112832090 | 0.32 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr12_+_44152740 | 0.32 |
ENST00000440781.2
ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4
|
interleukin-1 receptor-associated kinase 4 |
| chr15_+_67841330 | 0.31 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr11_-_58980342 | 0.31 |
ENST00000361050.3
|
MPEG1
|
macrophage expressed 1 |
| chr11_+_65101225 | 0.30 |
ENST00000528416.1
ENST00000415073.2 ENST00000252268.4 |
DPF2
|
D4, zinc and double PHD fingers family 2 |
| chr8_-_82359662 | 0.30 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr6_-_32821599 | 0.30 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr6_+_116850174 | 0.28 |
ENST00000416171.2
ENST00000368597.2 ENST00000452373.1 ENST00000405399.1 |
FAM26D
|
family with sequence similarity 26, member D |
| chr5_+_138609441 | 0.28 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
| chr15_+_85525205 | 0.28 |
ENST00000394553.1
ENST00000339708.5 |
PDE8A
|
phosphodiesterase 8A |
| chrX_-_50386648 | 0.27 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chr1_+_42619070 | 0.26 |
ENST00000372581.1
|
GUCA2B
|
guanylate cyclase activator 2B (uroguanylin) |
| chr7_+_112120908 | 0.26 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr4_-_116034979 | 0.26 |
ENST00000264363.2
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr1_+_12123414 | 0.25 |
ENST00000263932.2
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr3_-_57233966 | 0.24 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr6_-_11232891 | 0.24 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr10_+_98064085 | 0.23 |
ENST00000419175.1
ENST00000371174.2 |
DNTT
|
DNA nucleotidylexotransferase |
| chr17_+_34538310 | 0.23 |
ENST00000444414.1
ENST00000378350.4 ENST00000389068.5 ENST00000588929.1 ENST00000589079.1 ENST00000589336.1 ENST00000400702.4 ENST00000591167.1 ENST00000586598.1 ENST00000591637.1 ENST00000378352.4 ENST00000358756.5 |
CCL4L1
|
chemokine (C-C motif) ligand 4-like 1 |
| chr12_+_56435637 | 0.23 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr14_+_38677123 | 0.23 |
ENST00000267377.2
|
SSTR1
|
somatostatin receptor 1 |
| chr7_-_76829125 | 0.23 |
ENST00000248598.5
|
FGL2
|
fibrinogen-like 2 |
| chr17_-_62308087 | 0.23 |
ENST00000583097.1
|
TEX2
|
testis expressed 2 |
| chr9_+_131464767 | 0.22 |
ENST00000291906.4
|
PKN3
|
protein kinase N3 |
| chr5_+_147258266 | 0.22 |
ENST00000296694.4
|
SCGB3A2
|
secretoglobin, family 3A, member 2 |
| chr1_-_108735440 | 0.22 |
ENST00000370041.4
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr7_-_122840015 | 0.22 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr1_+_202431859 | 0.22 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr16_-_67694597 | 0.21 |
ENST00000393919.4
ENST00000219251.8 |
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr3_+_51575596 | 0.21 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr4_+_166300084 | 0.21 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr2_-_16804320 | 0.21 |
ENST00000355549.2
|
FAM49A
|
family with sequence similarity 49, member A |
| chr9_-_100000957 | 0.20 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr2_-_203735484 | 0.20 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr9_-_111619239 | 0.20 |
ENST00000374667.3
|
ACTL7B
|
actin-like 7B |
| chrX_+_102192200 | 0.19 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr2_+_103378472 | 0.19 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr3_+_121311966 | 0.18 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr14_-_46185155 | 0.18 |
ENST00000555442.1
|
RP11-369C8.1
|
RP11-369C8.1 |
| chr12_-_6451235 | 0.18 |
ENST00000440083.2
ENST00000162749.2 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr9_-_3469181 | 0.17 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr17_+_7461613 | 0.17 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr13_-_74708372 | 0.17 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr12_+_52282001 | 0.17 |
ENST00000340970.4
|
ANKRD33
|
ankyrin repeat domain 33 |
| chr9_+_90112590 | 0.16 |
ENST00000472284.1
|
DAPK1
|
death-associated protein kinase 1 |
| chr12_-_82152420 | 0.16 |
ENST00000552948.1
ENST00000548586.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr6_+_84222220 | 0.16 |
ENST00000369700.3
|
PRSS35
|
protease, serine, 35 |
| chr8_+_97506033 | 0.16 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr1_+_89246647 | 0.16 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr17_+_34640031 | 0.15 |
ENST00000339270.6
ENST00000482104.1 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr8_-_117886955 | 0.15 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr12_+_52281744 | 0.15 |
ENST00000301190.6
ENST00000538991.1 |
ANKRD33
|
ankyrin repeat domain 33 |
| chr2_+_234686976 | 0.15 |
ENST00000389758.3
|
MROH2A
|
maestro heat-like repeat family member 2A |
| chrX_-_100546314 | 0.14 |
ENST00000356784.1
|
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chr9_-_135996537 | 0.14 |
ENST00000372050.3
ENST00000372047.3 |
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr12_+_6493199 | 0.14 |
ENST00000228918.4
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr12_-_82153087 | 0.14 |
ENST00000547623.1
ENST00000549396.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr21_+_17792672 | 0.13 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_+_34639793 | 0.12 |
ENST00000394465.2
ENST00000394463.2 ENST00000378342.4 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr22_-_29457832 | 0.12 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr20_+_42136308 | 0.12 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr15_-_23932437 | 0.11 |
ENST00000331837.4
|
NDN
|
necdin, melanoma antigen (MAGE) family member |
| chr19_+_13229126 | 0.11 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr18_-_51107372 | 0.11 |
ENST00000565170.1
|
RP11-202D1.2
|
RP11-202D1.2 |
| chr11_+_63655987 | 0.11 |
ENST00000509502.2
ENST00000512060.1 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr5_+_150051149 | 0.11 |
ENST00000523553.1
|
MYOZ3
|
myozenin 3 |
| chr15_+_25200074 | 0.10 |
ENST00000390687.4
ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr5_-_88119580 | 0.10 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr12_+_80730292 | 0.10 |
ENST00000298820.3
|
OTOGL
|
otogelin-like |
| chr15_+_69857515 | 0.10 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr17_-_34890665 | 0.10 |
ENST00000586007.1
|
MYO19
|
myosin XIX |
| chr9_+_90112117 | 0.09 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr1_-_173572181 | 0.09 |
ENST00000536496.1
ENST00000367714.3 |
SLC9C2
|
solute carrier family 9, member C2 (putative) |
| chr2_-_204400013 | 0.09 |
ENST00000374489.2
ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr20_+_57209828 | 0.08 |
ENST00000598340.1
|
MGC4294
|
HCG1785561; MGC4294 protein; Uncharacterized protein |
| chr19_+_15052301 | 0.08 |
ENST00000248072.3
|
OR7C2
|
olfactory receptor, family 7, subfamily C, member 2 |
| chr1_+_152691998 | 0.08 |
ENST00000368775.2
|
C1orf68
|
chromosome 1 open reading frame 68 |
| chr17_+_7461849 | 0.07 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr11_-_8964580 | 0.07 |
ENST00000325884.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr7_-_75401513 | 0.07 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr17_+_7462031 | 0.07 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr3_+_141121164 | 0.06 |
ENST00000510338.1
ENST00000504673.1 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr17_+_7461781 | 0.06 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_+_234526272 | 0.06 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr15_+_91643173 | 0.06 |
ENST00000545111.2
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr9_+_90112741 | 0.06 |
ENST00000469640.2
|
DAPK1
|
death-associated protein kinase 1 |
| chr17_+_7461580 | 0.05 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr8_+_104311059 | 0.05 |
ENST00000358755.4
ENST00000523739.1 ENST00000540287.1 |
FZD6
|
frizzled family receptor 6 |
| chr7_-_15726296 | 0.05 |
ENST00000262041.5
|
MEOX2
|
mesenchyme homeobox 2 |
| chr10_-_61122220 | 0.05 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr4_+_71337834 | 0.04 |
ENST00000304887.5
|
MUC7
|
mucin 7, secreted |
| chr14_+_22520762 | 0.04 |
ENST00000390449.3
|
TRAV21
|
T cell receptor alpha variable 21 |
| chr2_-_175462456 | 0.03 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chrX_+_100878079 | 0.03 |
ENST00000471229.2
|
ARMCX3
|
armadillo repeat containing, X-linked 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.5 | 1.4 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.5 | 1.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 1.6 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.3 | 2.7 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.3 | 2.8 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.2 | 2.0 | GO:0030578 | PML body organization(GO:0030578) |
| 0.2 | 0.9 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 | 1.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 1.0 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.2 | 0.6 | GO:0043049 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.2 | 1.5 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.2 | 1.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 1.4 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.1 | 2.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.4 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.4 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 8.8 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 1.0 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 0.5 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.1 | 0.5 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.5 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.1 | 1.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 1.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.3 | GO:0032679 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.1 | 0.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.2 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 0.7 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.4 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.5 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 1.5 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.2 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.9 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.6 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 1.0 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.3 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 1.6 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 1.4 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.3 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 1.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.5 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.6 | GO:0043086 | negative regulation of catalytic activity(GO:0043086) |
| 0.0 | 0.3 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 0.7 | GO:0014047 | glutamate secretion(GO:0014047) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 0.8 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 2.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 11.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.6 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 1.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.6 | 2.8 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.5 | 1.6 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.5 | 2.5 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.3 | 1.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 1.0 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.2 | 2.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 1.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 1.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 1.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.7 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.4 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.4 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.5 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 1.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.5 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 2.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 2.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 9.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.3 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 1.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 1.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.4 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 2.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 3.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 4.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.8 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.1 | 2.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.9 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 1.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 2.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 3.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 2.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |