Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
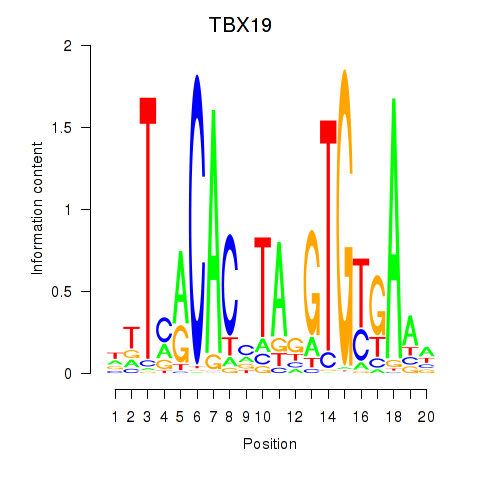
Results for TBX19
Z-value: 0.45
Transcription factors associated with TBX19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX19
|
ENSG00000143178.8 | T-box transcription factor 19 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX19 | hg19_v2_chr1_+_168250194_168250278 | 0.20 | 2.9e-01 | Click! |
Activity profile of TBX19 motif
Sorted Z-values of TBX19 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_7921090 | 2.48 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr7_+_154720173 | 1.01 |
ENST00000397551.2
|
PAXIP1-AS2
|
PAXIP1 antisense RNA 2 |
| chr3_+_98482175 | 0.99 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr3_+_94657086 | 0.89 |
ENST00000463200.1
|
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr19_+_58514229 | 0.88 |
ENST00000546949.1
ENST00000553254.1 ENST00000547364.1 |
CTD-2368P22.1
|
HCG1811579; Uncharacterized protein |
| chr2_-_113594279 | 0.78 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr17_-_39203519 | 0.66 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr2_+_143886877 | 0.58 |
ENST00000295095.6
|
ARHGAP15
|
Rho GTPase activating protein 15 |
| chr7_-_148580563 | 0.52 |
ENST00000476773.1
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr1_-_63988846 | 0.49 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr6_-_27880174 | 0.48 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr1_+_172389821 | 0.42 |
ENST00000367727.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr14_+_23727694 | 0.41 |
ENST00000399905.1
ENST00000470456.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr22_+_31477296 | 0.40 |
ENST00000426927.1
ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN
|
smoothelin |
| chr19_-_58514129 | 0.36 |
ENST00000552184.1
ENST00000546715.1 ENST00000536132.1 ENST00000547828.1 ENST00000547121.1 ENST00000551380.1 |
ZNF606
|
zinc finger protein 606 |
| chr12_-_9360966 | 0.35 |
ENST00000261336.2
|
PZP
|
pregnancy-zone protein |
| chr11_-_84028180 | 0.34 |
ENST00000280241.8
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr9_+_78505554 | 0.34 |
ENST00000545128.1
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr11_+_5474638 | 0.34 |
ENST00000341449.2
|
OR51I2
|
olfactory receptor, family 51, subfamily I, member 2 |
| chr9_+_78505581 | 0.33 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr15_+_26360970 | 0.30 |
ENST00000556159.1
ENST00000557523.1 |
LINC00929
|
long intergenic non-protein coding RNA 929 |
| chr17_+_3118915 | 0.30 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr12_-_49319265 | 0.30 |
ENST00000552878.1
ENST00000453172.2 |
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr20_+_43835638 | 0.29 |
ENST00000372781.3
ENST00000244069.6 |
SEMG1
|
semenogelin I |
| chr19_+_852291 | 0.28 |
ENST00000263621.1
|
ELANE
|
elastase, neutrophil expressed |
| chr7_-_122635754 | 0.28 |
ENST00000249284.2
|
TAS2R16
|
taste receptor, type 2, member 16 |
| chr1_-_243326612 | 0.27 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chrX_+_78426469 | 0.27 |
ENST00000276077.1
|
GPR174
|
G protein-coupled receptor 174 |
| chr17_-_56296580 | 0.27 |
ENST00000313863.6
ENST00000546108.1 ENST00000337050.7 ENST00000393119.2 |
MKS1
|
Meckel syndrome, type 1 |
| chr1_-_111174054 | 0.27 |
ENST00000369770.3
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr2_-_9563469 | 0.26 |
ENST00000484735.1
ENST00000456913.2 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr6_+_147091575 | 0.26 |
ENST00000326916.8
ENST00000470716.2 ENST00000367488.1 |
ADGB
|
androglobin |
| chr12_+_8276495 | 0.25 |
ENST00000546339.1
|
CLEC4A
|
C-type lectin domain family 4, member A |
| chr5_+_54320078 | 0.25 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr8_-_134115118 | 0.25 |
ENST00000395352.3
ENST00000338087.5 |
SLA
|
Src-like-adaptor |
| chr9_-_100684845 | 0.23 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr3_+_132316081 | 0.23 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr1_-_154458520 | 0.23 |
ENST00000486773.1
|
SHE
|
Src homology 2 domain containing E |
| chr19_+_7049332 | 0.23 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3-like 2 |
| chr5_-_39270725 | 0.22 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr11_-_11747257 | 0.22 |
ENST00000601641.1
|
AC131935.1
|
AC131935.1 |
| chrX_-_46187069 | 0.21 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr5_+_169010638 | 0.21 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr19_+_46003056 | 0.21 |
ENST00000401593.1
ENST00000396736.2 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr6_+_42847649 | 0.21 |
ENST00000424341.2
ENST00000602561.1 |
RPL7L1
|
ribosomal protein L7-like 1 |
| chr3_+_148447887 | 0.21 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr1_+_236557569 | 0.21 |
ENST00000334232.4
|
EDARADD
|
EDAR-associated death domain |
| chr7_+_100663353 | 0.21 |
ENST00000306151.4
|
MUC17
|
mucin 17, cell surface associated |
| chr10_+_12171636 | 0.20 |
ENST00000379051.1
ENST00000379033.3 ENST00000441368.1 ENST00000298428.9 ENST00000304267.8 |
SEC61A2
|
Sec61 alpha 2 subunit (S. cerevisiae) |
| chr15_+_84908573 | 0.20 |
ENST00000424966.1
ENST00000422563.2 |
GOLGA6L4
|
golgin A6 family-like 4 |
| chr6_+_26204825 | 0.20 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr20_-_22565101 | 0.20 |
ENST00000419308.2
|
FOXA2
|
forkhead box A2 |
| chr6_+_22569784 | 0.20 |
ENST00000510882.2
|
HDGFL1
|
hepatoma derived growth factor-like 1 |
| chrX_+_79675965 | 0.19 |
ENST00000308293.5
|
FAM46D
|
family with sequence similarity 46, member D |
| chr11_+_46740730 | 0.19 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr14_+_70346125 | 0.18 |
ENST00000361956.3
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr8_-_6795823 | 0.18 |
ENST00000297435.2
|
DEFA4
|
defensin, alpha 4, corticostatin |
| chr4_-_57976544 | 0.18 |
ENST00000295666.4
ENST00000537922.1 |
IGFBP7
|
insulin-like growth factor binding protein 7 |
| chr12_-_10978957 | 0.18 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr19_+_535835 | 0.18 |
ENST00000607527.1
ENST00000606065.1 |
CDC34
|
cell division cycle 34 |
| chr17_+_35306175 | 0.18 |
ENST00000225402.5
|
AATF
|
apoptosis antagonizing transcription factor |
| chr9_+_77230499 | 0.18 |
ENST00000396204.2
|
RORB
|
RAR-related orphan receptor B |
| chr12_-_11175219 | 0.18 |
ENST00000390673.2
|
TAS2R19
|
taste receptor, type 2, member 19 |
| chr2_+_62900986 | 0.18 |
ENST00000405015.3
ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1
|
EH domain binding protein 1 |
| chr6_-_10694766 | 0.17 |
ENST00000460742.2
ENST00000259983.3 ENST00000379586.1 |
C6orf52
|
chromosome 6 open reading frame 52 |
| chr8_+_94767072 | 0.17 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr18_+_61554932 | 0.17 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr9_+_125137565 | 0.17 |
ENST00000373698.5
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr11_-_19262486 | 0.17 |
ENST00000250024.4
|
E2F8
|
E2F transcription factor 8 |
| chr16_+_84801852 | 0.17 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr19_-_40331345 | 0.16 |
ENST00000597224.1
|
FBL
|
fibrillarin |
| chr6_-_26285737 | 0.16 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr6_+_96463840 | 0.16 |
ENST00000302103.5
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase) |
| chr12_-_8218997 | 0.16 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr7_-_142139783 | 0.16 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6 |
| chr1_+_117297007 | 0.15 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr20_+_44036620 | 0.15 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr12_+_120740119 | 0.15 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr5_-_33984741 | 0.15 |
ENST00000382102.3
ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2
|
solute carrier family 45, member 2 |
| chr5_-_177659761 | 0.14 |
ENST00000308158.5
|
PHYKPL
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr5_-_33984786 | 0.14 |
ENST00000296589.4
|
SLC45A2
|
solute carrier family 45, member 2 |
| chr6_-_38607673 | 0.14 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr3_+_119499331 | 0.14 |
ENST00000393716.2
ENST00000466380.1 |
NR1I2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr5_-_95768973 | 0.14 |
ENST00000311106.3
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1 |
| chr1_-_23520755 | 0.14 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr2_-_201753980 | 0.13 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr4_+_185570871 | 0.13 |
ENST00000512834.1
|
PRIMPOL
|
primase and polymerase (DNA-directed) |
| chr5_-_78281775 | 0.13 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr2_+_90153696 | 0.13 |
ENST00000417279.2
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr1_-_100598444 | 0.13 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chr2_-_201753717 | 0.13 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr10_+_71561630 | 0.13 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr2_-_201753859 | 0.13 |
ENST00000409361.1
ENST00000392283.4 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr19_+_49496705 | 0.13 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr19_+_49496782 | 0.12 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr16_-_2031464 | 0.12 |
ENST00000356120.4
ENST00000354249.4 |
NOXO1
|
NADPH oxidase organizer 1 |
| chr6_-_26216872 | 0.12 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr3_+_46204959 | 0.12 |
ENST00000357422.2
|
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr12_+_110011571 | 0.12 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr14_-_58894332 | 0.12 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr6_-_117086873 | 0.12 |
ENST00000368557.4
|
FAM162B
|
family with sequence similarity 162, member B |
| chr11_-_115375107 | 0.12 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr1_-_145382362 | 0.12 |
ENST00000419817.1
ENST00000421937.3 ENST00000433081.2 |
RP11-458D21.1
|
RP11-458D21.1 |
| chr6_-_31651817 | 0.12 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr8_+_94767109 | 0.12 |
ENST00000409623.3
ENST00000453906.1 ENST00000521517.1 |
TMEM67
|
transmembrane protein 67 |
| chr1_+_96457545 | 0.11 |
ENST00000413825.2
|
RP11-147C23.1
|
Uncharacterized protein |
| chr1_-_23521222 | 0.11 |
ENST00000374619.1
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr1_+_168250194 | 0.11 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr5_+_33440802 | 0.11 |
ENST00000502553.1
ENST00000514259.1 ENST00000265112.3 |
TARS
|
threonyl-tRNA synthetase |
| chr10_+_95653687 | 0.11 |
ENST00000371408.3
ENST00000427197.1 |
SLC35G1
|
solute carrier family 35, member G1 |
| chr1_+_100810575 | 0.10 |
ENST00000542213.1
|
CDC14A
|
cell division cycle 14A |
| chr2_-_64751227 | 0.10 |
ENST00000561559.1
|
RP11-568N6.1
|
RP11-568N6.1 |
| chr15_+_67835005 | 0.10 |
ENST00000178640.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr5_-_94417314 | 0.10 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr12_-_11036844 | 0.10 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr13_-_97646604 | 0.10 |
ENST00000298440.1
ENST00000543457.1 ENST00000541038.1 |
OXGR1
|
oxoglutarate (alpha-ketoglutarate) receptor 1 |
| chr1_+_160765947 | 0.09 |
ENST00000263285.6
ENST00000368039.2 |
LY9
|
lymphocyte antigen 9 |
| chr22_+_37678424 | 0.09 |
ENST00000248901.6
|
CYTH4
|
cytohesin 4 |
| chrX_+_150866779 | 0.09 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr1_-_116311402 | 0.09 |
ENST00000261448.5
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr7_+_2394445 | 0.09 |
ENST00000360876.4
ENST00000413917.1 ENST00000397011.2 |
EIF3B
|
eukaryotic translation initiation factor 3, subunit B |
| chr19_+_46732988 | 0.09 |
ENST00000437936.1
|
IGFL1
|
IGF-like family member 1 |
| chr7_+_134671234 | 0.08 |
ENST00000436302.2
ENST00000359383.3 ENST00000458078.1 ENST00000435976.2 ENST00000455283.2 |
AGBL3
|
ATP/GTP binding protein-like 3 |
| chr4_-_156298028 | 0.08 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr12_-_11214893 | 0.08 |
ENST00000533467.1
|
TAS2R46
|
taste receptor, type 2, member 46 |
| chr6_-_15586238 | 0.08 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr19_+_35168547 | 0.08 |
ENST00000502743.1
ENST00000509528.1 ENST00000506901.1 |
ZNF302
|
zinc finger protein 302 |
| chr5_+_140800638 | 0.08 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr9_+_130478345 | 0.08 |
ENST00000373289.3
ENST00000393748.4 |
TTC16
|
tetratricopeptide repeat domain 16 |
| chr5_-_110848189 | 0.08 |
ENST00000296632.3
ENST00000512160.1 ENST00000509887.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr2_+_120770645 | 0.07 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr6_-_131277510 | 0.07 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chrX_-_119709637 | 0.07 |
ENST00000404115.3
|
CUL4B
|
cullin 4B |
| chr2_-_162931052 | 0.07 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4 |
| chr3_+_35722487 | 0.07 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr6_+_26158343 | 0.07 |
ENST00000377777.4
ENST00000289316.2 |
HIST1H2BD
|
histone cluster 1, H2bd |
| chr3_+_2933893 | 0.07 |
ENST00000397459.2
|
CNTN4
|
contactin 4 |
| chr10_+_106034884 | 0.07 |
ENST00000369707.2
ENST00000429569.2 |
GSTO2
|
glutathione S-transferase omega 2 |
| chr2_-_217724767 | 0.06 |
ENST00000236979.2
|
TNP1
|
transition protein 1 (during histone to protamine replacement) |
| chrX_+_129535937 | 0.06 |
ENST00000305536.6
ENST00000370947.1 |
RBMX2
|
RNA binding motif protein, X-linked 2 |
| chr8_+_125954281 | 0.06 |
ENST00000510897.2
ENST00000533286.1 |
LINC00964
|
long intergenic non-protein coding RNA 964 |
| chr16_+_66995121 | 0.06 |
ENST00000303334.4
|
CES3
|
carboxylesterase 3 |
| chr1_-_150947343 | 0.05 |
ENST00000271688.6
ENST00000368954.5 |
CERS2
|
ceramide synthase 2 |
| chr16_+_66995144 | 0.05 |
ENST00000394037.1
|
CES3
|
carboxylesterase 3 |
| chr14_-_106573756 | 0.04 |
ENST00000390601.2
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr10_-_95241951 | 0.04 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr2_+_152214098 | 0.04 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr16_+_31404624 | 0.04 |
ENST00000389202.2
|
ITGAD
|
integrin, alpha D |
| chrX_+_107288239 | 0.04 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr10_-_95242044 | 0.04 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr19_+_21203426 | 0.04 |
ENST00000261560.5
ENST00000599548.1 ENST00000594110.1 |
ZNF430
|
zinc finger protein 430 |
| chr12_-_10962767 | 0.04 |
ENST00000240691.2
|
TAS2R9
|
taste receptor, type 2, member 9 |
| chr14_+_50159813 | 0.04 |
ENST00000359332.2
ENST00000553274.1 ENST00000557128.1 |
KLHDC1
|
kelch domain containing 1 |
| chr15_-_44010458 | 0.04 |
ENST00000541030.1
|
STRC
|
stereocilin |
| chr10_-_74714533 | 0.04 |
ENST00000373032.3
|
PLA2G12B
|
phospholipase A2, group XIIB |
| chr19_-_8008533 | 0.04 |
ENST00000597926.1
|
TIMM44
|
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chrX_+_107288197 | 0.04 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr11_-_5462744 | 0.03 |
ENST00000380211.1
|
OR51I1
|
olfactory receptor, family 51, subfamily I, member 1 |
| chr1_-_161039753 | 0.03 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr3_-_47555167 | 0.03 |
ENST00000296149.4
|
ELP6
|
elongator acetyltransferase complex subunit 6 |
| chr1_-_161039647 | 0.03 |
ENST00000368013.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr8_-_7343922 | 0.03 |
ENST00000335479.2
|
DEFB106B
|
defensin, beta 106B |
| chr8_-_19540086 | 0.03 |
ENST00000332246.6
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr17_+_36873677 | 0.03 |
ENST00000471200.1
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr22_-_31364187 | 0.03 |
ENST00000215862.4
ENST00000397641.3 |
MORC2
|
MORC family CW-type zinc finger 2 |
| chr8_-_19540266 | 0.02 |
ENST00000311540.4
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr1_+_16090914 | 0.02 |
ENST00000441801.2
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr22_-_47882857 | 0.02 |
ENST00000405369.3
|
LL22NC03-75H12.2
|
Novel protein; Uncharacterized protein |
| chr1_-_157567868 | 0.02 |
ENST00000271532.1
|
FCRL4
|
Fc receptor-like 4 |
| chr14_+_58894706 | 0.02 |
ENST00000261244.5
|
KIAA0586
|
KIAA0586 |
| chr9_-_130890662 | 0.02 |
ENST00000277462.5
ENST00000338961.6 |
PTGES2
|
prostaglandin E synthase 2 |
| chr12_-_10875831 | 0.02 |
ENST00000279550.7
ENST00000228251.4 |
YBX3
|
Y box binding protein 3 |
| chr10_-_12237820 | 0.02 |
ENST00000378937.3
ENST00000378927.3 |
NUDT5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr17_-_16118835 | 0.02 |
ENST00000582357.1
ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1
|
nuclear receptor corepressor 1 |
| chr4_+_71600144 | 0.02 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr4_+_123653807 | 0.02 |
ENST00000314218.3
ENST00000542236.1 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr10_+_76969909 | 0.02 |
ENST00000298468.5
ENST00000543351.1 |
VDAC2
|
voltage-dependent anion channel 2 |
| chr17_+_7123125 | 0.02 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr2_+_228337079 | 0.02 |
ENST00000409315.1
ENST00000373671.3 ENST00000409171.1 |
AGFG1
|
ArfGAP with FG repeats 1 |
| chr3_-_150264272 | 0.02 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr15_-_43910998 | 0.02 |
ENST00000450892.2
|
STRC
|
stereocilin |
| chr8_-_6836156 | 0.02 |
ENST00000382679.2
|
DEFA1
|
defensin, alpha 1 |
| chr4_-_47840112 | 0.02 |
ENST00000504584.1
|
CORIN
|
corin, serine peptidase |
| chr16_-_20339123 | 0.01 |
ENST00000381360.5
|
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chrX_+_119292467 | 0.01 |
ENST00000371388.3
|
RHOXF2
|
Rhox homeobox family, member 2 |
| chr19_-_39421377 | 0.01 |
ENST00000430193.3
ENST00000600042.1 ENST00000221431.6 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr16_+_3184924 | 0.01 |
ENST00000574902.1
ENST00000396878.3 |
ZNF213
|
zinc finger protein 213 |
| chr5_-_175461683 | 0.01 |
ENST00000514861.1
|
THOC3
|
THO complex 3 |
| chr3_+_185431080 | 0.01 |
ENST00000296270.1
|
C3orf65
|
chromosome 3 open reading frame 65 |
| chr12_+_8276224 | 0.01 |
ENST00000229332.5
|
CLEC4A
|
C-type lectin domain family 4, member A |
| chr10_+_12237924 | 0.01 |
ENST00000429258.2
ENST00000281141.4 |
CDC123
|
cell division cycle 123 |
| chr1_-_176176629 | 0.01 |
ENST00000367669.3
|
RFWD2
|
ring finger and WD repeat domain 2, E3 ubiquitin protein ligase |
| chr7_+_93535817 | 0.01 |
ENST00000248572.5
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr22_+_45072925 | 0.01 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr10_-_12237836 | 0.01 |
ENST00000444732.1
ENST00000378940.3 |
NUDT5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr22_+_45072958 | 0.01 |
ENST00000403581.1
|
PRR5
|
proline rich 5 (renal) |
| chr11_-_31531121 | 0.01 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr18_-_3874752 | 0.00 |
ENST00000534970.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr21_-_35884573 | 0.00 |
ENST00000399286.2
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chr14_+_105212297 | 0.00 |
ENST00000556623.1
ENST00000555674.1 |
ADSSL1
|
adenylosuccinate synthase like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX19
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.3 | 0.8 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.2 | 0.5 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.7 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.3 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.5 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.1 | 0.2 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.3 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.1 | 0.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.2 | GO:0034963 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 0.3 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.1 | 0.3 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0048736 | appendage development(GO:0048736) limb development(GO:0060173) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 1.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.2 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.3 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.5 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.5 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.2 | 1.0 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.3 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.2 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 1.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |