Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
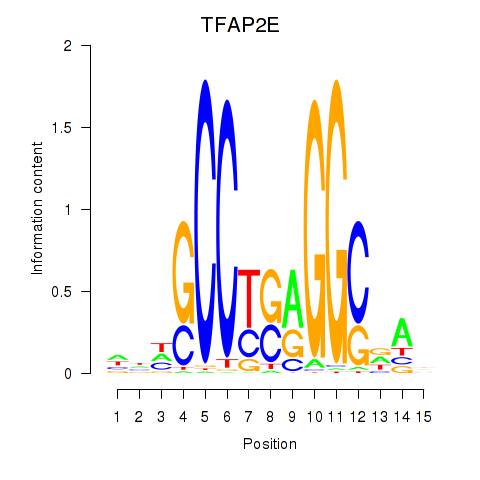
Results for TFAP2E
Z-value: 0.41
Transcription factors associated with TFAP2E
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2E
|
ENSG00000116819.6 | transcription factor AP-2 epsilon |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFAP2E | hg19_v2_chr1_+_36038971_36038971 | -0.38 | 4.1e-02 | Click! |
Activity profile of TFAP2E motif
Sorted Z-values of TFAP2E motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_65503801 | 1.09 |
ENST00000261883.4
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr19_+_45504688 | 0.94 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr2_+_38893208 | 0.85 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr19_-_35981358 | 0.64 |
ENST00000484218.2
ENST00000338897.3 |
KRTDAP
|
keratinocyte differentiation-associated protein |
| chr1_-_151778630 | 0.57 |
ENST00000368820.3
|
LINGO4
|
leucine rich repeat and Ig domain containing 4 |
| chr11_-_82612727 | 0.45 |
ENST00000531128.1
ENST00000535099.1 ENST00000527444.1 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr5_-_179499108 | 0.39 |
ENST00000521389.1
|
RNF130
|
ring finger protein 130 |
| chr19_-_54676884 | 0.39 |
ENST00000376591.4
|
TMC4
|
transmembrane channel-like 4 |
| chr5_-_179499086 | 0.38 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr19_-_54676846 | 0.37 |
ENST00000301187.4
|
TMC4
|
transmembrane channel-like 4 |
| chr12_-_15942503 | 0.37 |
ENST00000281172.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr17_+_72428218 | 0.35 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr14_-_25519095 | 0.33 |
ENST00000419632.2
ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr19_-_11039188 | 0.33 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr14_-_25519317 | 0.32 |
ENST00000323944.5
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr12_-_15942309 | 0.32 |
ENST00000544064.1
ENST00000543523.1 ENST00000536793.1 |
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr2_-_45236540 | 0.31 |
ENST00000303077.6
|
SIX2
|
SIX homeobox 2 |
| chr1_-_32801825 | 0.31 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr2_-_174828892 | 0.31 |
ENST00000418194.2
|
SP3
|
Sp3 transcription factor |
| chr1_+_211433275 | 0.29 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr9_-_86432547 | 0.28 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr2_+_38893047 | 0.26 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr2_+_232573208 | 0.26 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr20_+_55205825 | 0.24 |
ENST00000544508.1
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chrX_+_128872998 | 0.23 |
ENST00000371106.3
|
XPNPEP2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr2_+_232573222 | 0.23 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chrX_-_124097620 | 0.23 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr17_-_39769005 | 0.21 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr1_+_17634689 | 0.20 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr17_+_37824700 | 0.20 |
ENST00000581428.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr3_+_49591881 | 0.18 |
ENST00000296452.4
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr16_+_69599861 | 0.17 |
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr16_+_69599899 | 0.17 |
ENST00000567239.1
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr3_+_32280159 | 0.17 |
ENST00000458535.2
ENST00000307526.3 |
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr16_+_69600058 | 0.16 |
ENST00000393742.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr22_+_31608219 | 0.16 |
ENST00000406516.1
ENST00000444929.2 ENST00000331728.4 |
LIMK2
|
LIM domain kinase 2 |
| chr3_-_136471204 | 0.16 |
ENST00000480733.1
ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1
|
stromal antigen 1 |
| chr11_-_118305921 | 0.15 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr17_-_42992856 | 0.15 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr20_+_36888551 | 0.14 |
ENST00000418004.1
ENST00000451435.1 |
BPI
|
bactericidal/permeability-increasing protein |
| chr3_+_105086056 | 0.14 |
ENST00000472644.2
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr12_+_75784850 | 0.14 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr11_+_64009072 | 0.14 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr1_+_152943122 | 0.14 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chrX_+_152990302 | 0.13 |
ENST00000218104.3
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr1_-_201346761 | 0.13 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr19_+_36195429 | 0.13 |
ENST00000392197.2
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chrX_+_51636629 | 0.12 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr10_-_112678976 | 0.12 |
ENST00000448814.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr12_-_65153175 | 0.12 |
ENST00000543646.1
ENST00000542058.1 ENST00000258145.3 |
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr12_+_53773944 | 0.11 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr17_-_47755436 | 0.11 |
ENST00000505581.1
ENST00000514121.1 ENST00000393328.2 ENST00000509079.1 ENST00000393331.3 ENST00000347630.2 ENST00000504102.1 |
SPOP
|
speckle-type POZ protein |
| chr10_-_98480243 | 0.11 |
ENST00000339364.5
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr16_-_28506840 | 0.11 |
ENST00000569430.1
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr17_+_48423453 | 0.11 |
ENST00000017003.2
ENST00000509778.1 ENST00000507602.1 |
XYLT2
|
xylosyltransferase II |
| chr1_+_233749739 | 0.11 |
ENST00000366621.3
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr22_+_31644388 | 0.10 |
ENST00000333611.4
ENST00000340552.4 |
LIMK2
|
LIM domain kinase 2 |
| chr1_+_9599540 | 0.10 |
ENST00000302692.6
|
SLC25A33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chrX_+_152783131 | 0.10 |
ENST00000349466.2
ENST00000370186.1 |
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr9_+_19230433 | 0.10 |
ENST00000434457.2
ENST00000602925.1 |
DENND4C
|
DENN/MADD domain containing 4C |
| chr8_+_146277764 | 0.10 |
ENST00000331434.6
|
C8orf33
|
chromosome 8 open reading frame 33 |
| chr5_-_179498703 | 0.10 |
ENST00000522208.2
|
RNF130
|
ring finger protein 130 |
| chr19_-_46389359 | 0.10 |
ENST00000302165.3
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1 |
| chr3_+_4535025 | 0.09 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr19_-_11039261 | 0.09 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr1_+_35225339 | 0.09 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr2_-_24149977 | 0.09 |
ENST00000238789.5
|
ATAD2B
|
ATPase family, AAA domain containing 2B |
| chr11_+_63974135 | 0.09 |
ENST00000544997.1
ENST00000345728.5 ENST00000279227.5 |
FERMT3
|
fermitin family member 3 |
| chr19_+_50936142 | 0.08 |
ENST00000357701.5
|
MYBPC2
|
myosin binding protein C, fast type |
| chr3_-_55521323 | 0.08 |
ENST00000264634.4
|
WNT5A
|
wingless-type MMTV integration site family, member 5A |
| chr19_+_36195467 | 0.08 |
ENST00000426659.2
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr10_+_99894380 | 0.08 |
ENST00000370584.3
|
R3HCC1L
|
R3H domain and coiled-coil containing 1-like |
| chr7_+_100464760 | 0.08 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr20_+_58508817 | 0.08 |
ENST00000358293.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr3_+_110790590 | 0.08 |
ENST00000485303.1
|
PVRL3
|
poliovirus receptor-related 3 |
| chr8_+_125463048 | 0.07 |
ENST00000328599.3
|
TRMT12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr1_-_154531095 | 0.07 |
ENST00000292211.4
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr3_+_52813932 | 0.07 |
ENST00000537050.1
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr10_-_112678692 | 0.07 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr5_-_179719048 | 0.07 |
ENST00000523583.1
ENST00000393360.3 ENST00000455781.1 ENST00000347470.4 ENST00000452135.2 ENST00000343111.6 |
MAPK9
|
mitogen-activated protein kinase 9 |
| chrX_-_118826784 | 0.07 |
ENST00000394616.4
|
SEPT6
|
septin 6 |
| chr5_+_176237478 | 0.06 |
ENST00000329542.4
|
UNC5A
|
unc-5 homolog A (C. elegans) |
| chr3_+_155860751 | 0.06 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr1_+_901847 | 0.06 |
ENST00000379410.3
ENST00000379409.2 ENST00000379407.3 |
PLEKHN1
|
pleckstrin homology domain containing, family N member 1 |
| chr19_+_3572758 | 0.05 |
ENST00000416526.1
|
HMG20B
|
high mobility group 20B |
| chr1_+_9005917 | 0.05 |
ENST00000549778.1
ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6
|
carbonic anhydrase VI |
| chr1_+_104068562 | 0.05 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr5_+_133984462 | 0.05 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr10_-_112678904 | 0.05 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr12_-_48213568 | 0.04 |
ENST00000080059.7
ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7
|
histone deacetylase 7 |
| chr1_+_150122034 | 0.04 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr4_+_41992489 | 0.04 |
ENST00000264451.7
|
SLC30A9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr20_-_30539773 | 0.04 |
ENST00000202017.4
|
PDRG1
|
p53 and DNA-damage regulated 1 |
| chr17_+_37824411 | 0.04 |
ENST00000269582.2
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr17_-_47755338 | 0.04 |
ENST00000508805.1
ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP
|
speckle-type POZ protein |
| chr8_+_17780346 | 0.04 |
ENST00000325083.8
|
PCM1
|
pericentriolar material 1 |
| chr11_-_66056596 | 0.04 |
ENST00000471387.2
ENST00000359461.6 ENST00000376901.4 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr1_+_104068312 | 0.04 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr10_+_99894399 | 0.04 |
ENST00000298999.3
ENST00000314594.5 |
R3HCC1L
|
R3H domain and coiled-coil containing 1-like |
| chr19_-_43969796 | 0.04 |
ENST00000244333.3
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr11_-_47869865 | 0.03 |
ENST00000530326.1
ENST00000532747.1 |
NUP160
|
nucleoporin 160kDa |
| chr20_-_13619550 | 0.03 |
ENST00000455532.1
ENST00000544472.1 ENST00000539805.1 ENST00000337743.4 |
TASP1
|
taspase, threonine aspartase, 1 |
| chr2_+_232572361 | 0.03 |
ENST00000409321.1
|
PTMA
|
prothymosin, alpha |
| chr17_+_37824217 | 0.03 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr11_-_47870019 | 0.03 |
ENST00000378460.2
|
NUP160
|
nucleoporin 160kDa |
| chr1_+_43148625 | 0.02 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr9_+_128024067 | 0.02 |
ENST00000461379.1
ENST00000394084.1 ENST00000394105.2 ENST00000470056.1 ENST00000394104.2 ENST00000265956.4 ENST00000394083.2 ENST00000495955.1 ENST00000467750.1 ENST00000297933.6 |
GAPVD1
|
GTPase activating protein and VPS9 domains 1 |
| chr1_+_182584314 | 0.02 |
ENST00000566297.1
|
RP11-317P15.4
|
RP11-317P15.4 |
| chr1_-_114355083 | 0.02 |
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1 |
| chr19_+_3572925 | 0.02 |
ENST00000333651.6
ENST00000417382.1 ENST00000453933.1 ENST00000262949.7 |
HMG20B
|
high mobility group 20B |
| chr6_+_37787262 | 0.01 |
ENST00000287218.4
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr13_-_76056250 | 0.01 |
ENST00000377636.3
ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4
|
TBC1 domain family, member 4 |
| chr6_+_42531798 | 0.01 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr14_-_64010046 | 0.01 |
ENST00000337537.3
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr3_+_51428704 | 0.01 |
ENST00000323686.4
|
RBM15B
|
RNA binding motif protein 15B |
| chr2_-_241831424 | 0.01 |
ENST00000402775.2
ENST00000307486.8 |
C2orf54
|
chromosome 2 open reading frame 54 |
| chr20_-_25371412 | 0.01 |
ENST00000339157.5
ENST00000376542.3 |
ABHD12
|
abhydrolase domain containing 12 |
| chr10_-_108924284 | 0.01 |
ENST00000344440.6
ENST00000263054.6 |
SORCS1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr19_+_11039391 | 0.01 |
ENST00000270502.6
|
C19orf52
|
chromosome 19 open reading frame 52 |
| chr22_+_35653445 | 0.01 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr3_+_4535155 | 0.01 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr1_-_231175964 | 0.00 |
ENST00000366654.4
|
FAM89A
|
family with sequence similarity 89, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP2E
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.7 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.5 | GO:0002254 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 1.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.3 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.1 | 0.9 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.6 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0061346 | cardiac right atrium morphogenesis(GO:0003213) negative regulation of melanin biosynthetic process(GO:0048022) positive regulation of anagen(GO:0051885) mediolateral intercalation(GO:0060031) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) negative regulation of secondary metabolite biosynthetic process(GO:1900377) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.3 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.1 | 1.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |