Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
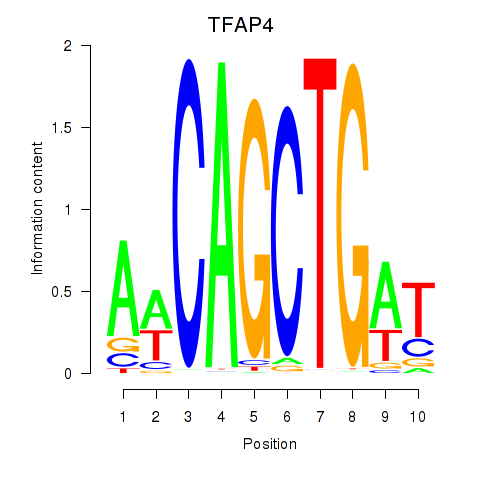
Results for TFAP4_MSC
Z-value: 0.71
Transcription factors associated with TFAP4_MSC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP4
|
ENSG00000090447.7 | transcription factor AP-4 |
|
MSC
|
ENSG00000178860.8 | musculin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSC | hg19_v2_chr8_-_72756667_72756736 | -0.32 | 8.9e-02 | Click! |
| TFAP4 | hg19_v2_chr16_-_4323015_4323076 | 0.16 | 3.9e-01 | Click! |
Activity profile of TFAP4_MSC motif
Sorted Z-values of TFAP4_MSC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_24911195 | 6.31 |
ENST00000259698.4
|
FAM65B
|
family with sequence similarity 65, member B |
| chr16_-_21289627 | 4.73 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr3_+_3841108 | 4.41 |
ENST00000319331.3
|
LRRN1
|
leucine rich repeat neuronal 1 |
| chr16_-_67427389 | 4.16 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr6_-_39197226 | 2.85 |
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5 |
| chr8_-_72274095 | 2.80 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr20_-_39317868 | 2.77 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr15_-_40401062 | 2.51 |
ENST00000354670.4
ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF
|
Bcl2 modifying factor |
| chr11_+_45918092 | 2.38 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr2_+_85981008 | 2.27 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr6_+_17281573 | 2.24 |
ENST00000379052.5
|
RBM24
|
RNA binding motif protein 24 |
| chr8_-_110660999 | 2.13 |
ENST00000424158.2
ENST00000533895.1 ENST00000446070.2 ENST00000528331.1 ENST00000526302.1 ENST00000433638.1 ENST00000408908.2 ENST00000524720.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr4_+_75858318 | 1.89 |
ENST00000307428.7
|
PARM1
|
prostate androgen-regulated mucin-like protein 1 |
| chr22_-_36013368 | 1.80 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr12_+_49297899 | 1.77 |
ENST00000552942.1
ENST00000320516.4 |
CCDC65
|
coiled-coil domain containing 65 |
| chr9_-_112970436 | 1.70 |
ENST00000400613.4
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr8_+_81398444 | 1.67 |
ENST00000455036.3
ENST00000426744.2 |
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr1_-_216896780 | 1.61 |
ENST00000459955.1
ENST00000366937.1 ENST00000408911.3 ENST00000391890.3 |
ESRRG
|
estrogen-related receptor gamma |
| chr4_+_75858290 | 1.59 |
ENST00000513238.1
|
PARM1
|
prostate androgen-regulated mucin-like protein 1 |
| chr5_+_50678921 | 1.55 |
ENST00000230658.7
|
ISL1
|
ISL LIM homeobox 1 |
| chr1_+_212738676 | 1.54 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr8_-_72274467 | 1.49 |
ENST00000340726.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr17_+_4981535 | 1.49 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr10_-_102089729 | 1.48 |
ENST00000465680.2
|
PKD2L1
|
polycystic kidney disease 2-like 1 |
| chr10_+_94594351 | 1.43 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr8_+_81397876 | 1.41 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr12_+_104458235 | 1.40 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr22_+_31003133 | 1.39 |
ENST00000405742.3
|
TCN2
|
transcobalamin II |
| chr1_-_183622442 | 1.36 |
ENST00000308641.4
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr22_+_31002779 | 1.36 |
ENST00000215838.3
|
TCN2
|
transcobalamin II |
| chr2_+_233562015 | 1.33 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr1_-_150669500 | 1.28 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr21_-_34185944 | 1.27 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr17_+_68071389 | 1.25 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr17_+_68071458 | 1.24 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr15_+_67547163 | 1.23 |
ENST00000335894.4
|
IQCH
|
IQ motif containing H |
| chr4_+_74702214 | 1.21 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr11_-_85430204 | 1.19 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr3_-_183273477 | 1.16 |
ENST00000341319.3
|
KLHL6
|
kelch-like family member 6 |
| chr11_-_85430163 | 1.15 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr1_+_14075865 | 1.11 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr11_+_73675873 | 1.08 |
ENST00000537753.1
ENST00000542350.1 |
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr16_+_83932684 | 1.07 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr7_-_120497178 | 1.01 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr3_+_49057876 | 1.00 |
ENST00000326912.4
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr12_+_56473628 | 0.94 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr3_+_49058444 | 0.93 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr1_-_39339777 | 0.93 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr12_+_49297887 | 0.92 |
ENST00000266984.5
|
CCDC65
|
coiled-coil domain containing 65 |
| chr11_+_7597639 | 0.91 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr22_+_35776828 | 0.91 |
ENST00000216117.8
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr7_-_30029574 | 0.90 |
ENST00000426154.1
ENST00000421434.1 ENST00000434476.2 |
SCRN1
|
secernin 1 |
| chr22_+_31003190 | 0.86 |
ENST00000407817.3
|
TCN2
|
transcobalamin II |
| chr4_+_86699834 | 0.85 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr2_-_73460334 | 0.84 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr7_+_128784712 | 0.84 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr7_-_30029367 | 0.82 |
ENST00000242059.5
|
SCRN1
|
secernin 1 |
| chr1_+_15272271 | 0.82 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr15_+_43809797 | 0.82 |
ENST00000399453.1
ENST00000300231.5 |
MAP1A
|
microtubule-associated protein 1A |
| chr1_+_110082487 | 0.81 |
ENST00000527748.1
|
GPR61
|
G protein-coupled receptor 61 |
| chr19_+_18208603 | 0.80 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr13_-_41635512 | 0.79 |
ENST00000405737.2
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr3_-_120365866 | 0.79 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr2_+_97203082 | 0.79 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr16_-_31076332 | 0.78 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr1_+_14075903 | 0.76 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr2_-_27603582 | 0.76 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr14_-_53019211 | 0.76 |
ENST00000557374.1
ENST00000281741.4 |
TXNDC16
|
thioredoxin domain containing 16 |
| chr5_+_102201687 | 0.76 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_-_24741525 | 0.76 |
ENST00000374409.1
|
STPG1
|
sperm-tail PG-rich repeat containing 1 |
| chr6_-_76203454 | 0.74 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr3_-_65583561 | 0.74 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_+_167063282 | 0.73 |
ENST00000361200.2
|
DUSP27
|
dual specificity phosphatase 27 (putative) |
| chr6_-_76203345 | 0.73 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr2_+_230787201 | 0.73 |
ENST00000283946.3
|
FBXO36
|
F-box protein 36 |
| chr7_-_99869799 | 0.73 |
ENST00000436886.2
|
GATS
|
GATS, stromal antigen 3 opposite strand |
| chr12_+_109826524 | 0.72 |
ENST00000431443.2
|
MYO1H
|
myosin IH |
| chr7_+_116660246 | 0.72 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr11_-_62474803 | 0.72 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr5_+_102201509 | 0.71 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr10_+_95848824 | 0.71 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chrX_-_10851762 | 0.70 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr3_-_52486841 | 0.70 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr20_+_44098346 | 0.69 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr12_+_72233487 | 0.69 |
ENST00000482439.2
ENST00000550746.1 ENST00000491063.1 ENST00000319106.8 ENST00000485960.2 ENST00000393309.3 |
TBC1D15
|
TBC1 domain family, member 15 |
| chr6_-_31697255 | 0.69 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr6_+_46761118 | 0.68 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr5_+_102201722 | 0.67 |
ENST00000274392.9
ENST00000455264.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_-_158345462 | 0.66 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr12_+_122150646 | 0.66 |
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B |
| chr3_-_64211112 | 0.64 |
ENST00000295902.6
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr6_+_44184653 | 0.63 |
ENST00000573382.2
ENST00000576476.1 |
RP1-302G2.5
|
RP1-302G2.5 |
| chr6_+_90272027 | 0.63 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr19_+_10527449 | 0.62 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr14_-_23288930 | 0.62 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr17_+_58755184 | 0.62 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr12_-_54121261 | 0.61 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr11_+_71498552 | 0.61 |
ENST00000346333.6
ENST00000359244.4 ENST00000426628.2 |
FAM86C1
|
family with sequence similarity 86, member C1 |
| chr3_-_193272741 | 0.61 |
ENST00000392443.3
|
ATP13A4
|
ATPase type 13A4 |
| chr3_-_193272874 | 0.59 |
ENST00000342695.4
|
ATP13A4
|
ATPase type 13A4 |
| chr11_+_1944054 | 0.59 |
ENST00000397301.1
ENST00000397304.2 ENST00000446240.1 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr1_-_86043921 | 0.59 |
ENST00000535924.2
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr12_-_54121212 | 0.58 |
ENST00000548263.1
ENST00000430117.2 ENST00000550804.1 ENST00000549173.1 ENST00000551900.1 ENST00000546619.1 ENST00000548177.1 ENST00000549349.1 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr12_+_86268065 | 0.58 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr2_+_95537170 | 0.57 |
ENST00000295201.4
|
TEKT4
|
tektin 4 |
| chr20_+_53092123 | 0.56 |
ENST00000262593.5
|
DOK5
|
docking protein 5 |
| chr22_+_39052632 | 0.55 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr1_-_175712665 | 0.55 |
ENST00000263525.2
|
TNR
|
tenascin R |
| chr8_-_95961578 | 0.55 |
ENST00000448464.2
ENST00000342697.4 |
TP53INP1
|
tumor protein p53 inducible nuclear protein 1 |
| chr16_-_31076273 | 0.54 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr20_+_33292068 | 0.54 |
ENST00000374810.3
ENST00000374809.2 ENST00000451665.1 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr14_+_45431379 | 0.54 |
ENST00000361577.3
ENST00000361462.2 ENST00000382233.2 |
FAM179B
|
family with sequence similarity 179, member B |
| chrX_-_108868390 | 0.54 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr8_+_98788003 | 0.54 |
ENST00000521545.2
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr1_+_174844645 | 0.53 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr16_-_4323015 | 0.53 |
ENST00000204517.6
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr2_-_148779106 | 0.52 |
ENST00000416719.1
ENST00000264169.2 |
ORC4
|
origin recognition complex, subunit 4 |
| chr8_+_98788057 | 0.52 |
ENST00000517924.1
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr3_-_52868931 | 0.52 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr15_-_75748115 | 0.51 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr20_-_40247133 | 0.51 |
ENST00000373233.3
ENST00000309279.7 |
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr12_-_50297638 | 0.51 |
ENST00000320634.3
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr20_+_53092232 | 0.50 |
ENST00000395939.1
|
DOK5
|
docking protein 5 |
| chr17_+_26800648 | 0.50 |
ENST00000545060.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr4_-_103682145 | 0.50 |
ENST00000226578.4
|
MANBA
|
mannosidase, beta A, lysosomal |
| chr17_+_26800756 | 0.49 |
ENST00000537681.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr20_+_43160409 | 0.49 |
ENST00000372894.3
ENST00000372892.3 ENST00000372891.3 |
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr9_-_123239632 | 0.49 |
ENST00000416449.1
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr2_-_154335300 | 0.49 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr8_-_71581377 | 0.49 |
ENST00000276590.4
ENST00000522447.1 |
LACTB2
|
lactamase, beta 2 |
| chr1_+_223101757 | 0.48 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr11_+_6411670 | 0.48 |
ENST00000530395.1
ENST00000527275.1 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr14_+_24584508 | 0.47 |
ENST00000559354.1
ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr5_+_102201430 | 0.47 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_+_172422026 | 0.47 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr15_-_52944231 | 0.47 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr17_+_1627834 | 0.46 |
ENST00000419248.1
ENST00000418841.1 |
WDR81
|
WD repeat domain 81 |
| chr5_+_49962772 | 0.46 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr6_-_31651817 | 0.46 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr18_-_19180681 | 0.45 |
ENST00000269214.5
|
ESCO1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr17_+_7462103 | 0.45 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr3_-_52869205 | 0.45 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr20_+_44098385 | 0.45 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr7_-_92855762 | 0.44 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr9_-_13165457 | 0.44 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr6_-_39399087 | 0.44 |
ENST00000229913.5
ENST00000541946.1 ENST00000394362.1 |
KIF6
|
kinesin family member 6 |
| chr12_-_50298000 | 0.43 |
ENST00000550635.2
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr16_-_5147743 | 0.43 |
ENST00000587133.1
ENST00000458008.4 ENST00000427587.4 |
FAM86A
|
family with sequence similarity 86, member A |
| chr17_+_26800296 | 0.43 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr5_+_43603229 | 0.43 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr17_+_7462031 | 0.43 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr17_-_47755436 | 0.42 |
ENST00000505581.1
ENST00000514121.1 ENST00000393328.2 ENST00000509079.1 ENST00000393331.3 ENST00000347630.2 ENST00000504102.1 |
SPOP
|
speckle-type POZ protein |
| chr2_+_231280954 | 0.42 |
ENST00000409824.1
ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100
|
SP100 nuclear antigen |
| chr3_-_49058479 | 0.42 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr1_+_159750720 | 0.42 |
ENST00000368109.1
ENST00000368108.3 |
DUSP23
|
dual specificity phosphatase 23 |
| chr2_+_68384976 | 0.42 |
ENST00000263657.2
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae) |
| chr1_+_159750776 | 0.42 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr11_+_6411636 | 0.41 |
ENST00000299397.3
ENST00000356761.2 ENST00000342245.4 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chrY_+_14774265 | 0.41 |
ENST00000457658.1
ENST00000440408.1 ENST00000543097.1 |
TTTY15
|
testis-specific transcript, Y-linked 15 (non-protein coding) |
| chr15_+_67547113 | 0.40 |
ENST00000512104.1
ENST00000358767.3 ENST00000546225.1 |
IQCH
|
IQ motif containing H |
| chr12_+_122667658 | 0.40 |
ENST00000339777.4
ENST00000425921.1 |
LRRC43
|
leucine rich repeat containing 43 |
| chr17_-_7145106 | 0.40 |
ENST00000577035.1
|
GABARAP
|
GABA(A) receptor-associated protein |
| chr5_+_145583156 | 0.40 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr9_+_2717502 | 0.40 |
ENST00000382082.3
|
KCNV2
|
potassium channel, subfamily V, member 2 |
| chr11_+_71938925 | 0.40 |
ENST00000538751.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr17_-_26989136 | 0.40 |
ENST00000247020.4
|
SDF2
|
stromal cell-derived factor 2 |
| chr10_-_62149433 | 0.39 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr19_-_46285646 | 0.39 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr9_+_2159850 | 0.39 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr11_-_85430088 | 0.39 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr20_+_62697564 | 0.39 |
ENST00000458442.1
|
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr5_-_58571935 | 0.38 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr2_+_137523086 | 0.38 |
ENST00000409968.1
|
THSD7B
|
thrombospondin, type I, domain containing 7B |
| chr12_-_56122426 | 0.38 |
ENST00000551173.1
|
CD63
|
CD63 molecule |
| chr6_-_31697563 | 0.38 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr16_+_71929397 | 0.38 |
ENST00000537613.1
ENST00000424485.2 ENST00000606369.1 ENST00000329908.8 ENST00000538850.1 ENST00000541918.1 ENST00000534994.1 ENST00000378798.5 ENST00000539186.1 |
IST1
|
increased sodium tolerance 1 homolog (yeast) |
| chr15_-_42749711 | 0.37 |
ENST00000565611.1
ENST00000263805.4 ENST00000565948.1 |
ZNF106
|
zinc finger protein 106 |
| chr2_+_97202480 | 0.37 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr15_-_83680325 | 0.37 |
ENST00000508990.2
ENST00000510873.2 ENST00000538348.2 ENST00000451195.3 ENST00000513601.2 ENST00000304177.5 ENST00000565712.1 |
C15orf40
|
chromosome 15 open reading frame 40 |
| chr16_-_84273304 | 0.37 |
ENST00000308251.4
ENST00000568181.1 |
KCNG4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr2_-_148778323 | 0.36 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr19_+_1041212 | 0.36 |
ENST00000433129.1
|
ABCA7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr19_+_7587491 | 0.36 |
ENST00000264079.6
|
MCOLN1
|
mucolipin 1 |
| chr1_-_33647267 | 0.36 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr8_-_38008783 | 0.35 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr10_+_88428206 | 0.35 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr17_-_38083843 | 0.35 |
ENST00000304046.2
ENST00000579695.1 |
ORMDL3
|
ORM1-like 3 (S. cerevisiae) |
| chr4_+_159727272 | 0.35 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr4_+_154178520 | 0.35 |
ENST00000433687.1
|
TRIM2
|
tripartite motif containing 2 |
| chr3_+_32147997 | 0.34 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr10_+_102790980 | 0.34 |
ENST00000393459.1
ENST00000224807.5 |
SFXN3
|
sideroflexin 3 |
| chr4_-_140223670 | 0.34 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr8_-_74791051 | 0.34 |
ENST00000453587.2
ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr6_+_30852130 | 0.33 |
ENST00000428153.2
ENST00000376568.3 ENST00000452441.1 ENST00000515219.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_+_179149636 | 0.33 |
ENST00000409631.1
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr6_+_30851840 | 0.32 |
ENST00000511510.1
ENST00000376569.3 ENST00000376575.3 ENST00000376570.4 ENST00000446312.1 ENST00000504927.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr17_+_73521763 | 0.32 |
ENST00000167462.5
ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr17_-_80275466 | 0.31 |
ENST00000312648.3
|
CD7
|
CD7 molecule |
| chr1_-_68915610 | 0.31 |
ENST00000262340.5
|
RPE65
|
retinal pigment epithelium-specific protein 65kDa |
| chr21_-_34185989 | 0.31 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP4_MSC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0035284 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.7 | 2.6 | GO:0018032 | protein amidation(GO:0018032) |
| 0.5 | 1.6 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) |
| 0.5 | 1.5 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.5 | 4.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 3.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.3 | 4.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.3 | 0.9 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.3 | 2.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 1.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 1.8 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.2 | 1.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 1.1 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 2.7 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.2 | 0.5 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.2 | 0.7 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.7 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.1 | 0.7 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 | 2.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.5 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.9 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 1.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.4 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.1 | 1.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.4 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.4 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 2.5 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 0.8 | GO:0051604 | protein maturation(GO:0051604) |
| 0.1 | 0.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.4 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 1.9 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 2.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 0.4 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.3 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.1 | 0.2 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.1 | 0.4 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.4 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.3 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.1 | 0.8 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.5 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.9 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.3 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.1 | 0.2 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 1.1 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 0.2 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 3.2 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.2 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.1 | 0.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.2 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 1.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.7 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.4 | GO:0032868 | response to insulin(GO:0032868) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 4.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 4.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 1.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.5 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.9 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.5 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.3 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.3 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.5 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.4 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.8 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 2.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 2.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.1 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 1.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.4 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.2 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0098727 | stem cell population maintenance(GO:0019827) maintenance of cell number(GO:0098727) |
| 0.0 | 0.2 | GO:0044598 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 1.6 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 1.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0070997 | neuron death(GO:0070997) |
| 0.0 | 0.0 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 1.0 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.3 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.2 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 0.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.4 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.0 | GO:0002265 | astrocyte activation involved in immune response(GO:0002265) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 1.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.4 | 4.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 2.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 0.7 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.2 | 1.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 2.1 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 6.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 1.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.9 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 4.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 3.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 2.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.6 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) junctional membrane complex(GO:0030314) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0044455 | mitochondrial membrane part(GO:0044455) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 2.0 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 3.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.7 | 2.6 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.5 | 1.4 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.3 | 3.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 0.9 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 1.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 1.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 0.6 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.2 | 2.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 1.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.6 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.4 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 2.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.4 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 2.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 1.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 2.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.4 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.3 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 2.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.3 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 1.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 1.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.4 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 2.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 1.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.7 | GO:0015923 | mannose binding(GO:0005537) mannosidase activity(GO:0015923) |
| 0.0 | 5.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 1.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 1.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 1.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 5.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.0 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.0 | 0.5 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.6 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.8 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 2.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 2.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |