Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
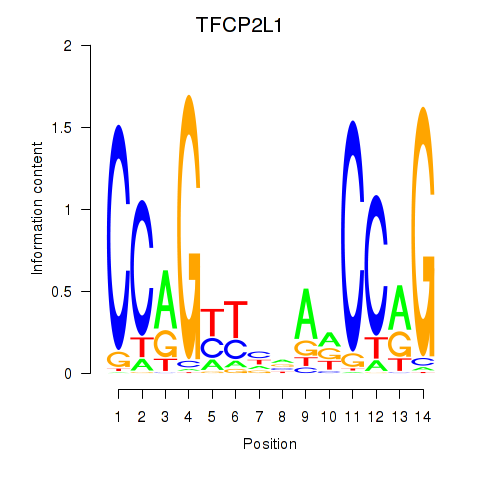
Results for TFCP2L1
Z-value: 0.59
Transcription factors associated with TFCP2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2L1
|
ENSG00000115112.7 | transcription factor CP2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2L1 | hg19_v2_chr2_-_122042770_122042785 | 0.17 | 3.6e-01 | Click! |
Activity profile of TFCP2L1 motif
Sorted Z-values of TFCP2L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_180018540 | 3.85 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr1_-_46089718 | 1.76 |
ENST00000421127.2
ENST00000343901.2 ENST00000528266.1 |
CCDC17
|
coiled-coil domain containing 17 |
| chr1_-_46089639 | 1.59 |
ENST00000445048.2
|
CCDC17
|
coiled-coil domain containing 17 |
| chr17_-_1532106 | 1.53 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr18_-_24765248 | 1.26 |
ENST00000580774.1
ENST00000284224.8 |
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr1_-_204329013 | 1.09 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr19_+_35629702 | 1.06 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr14_-_106069247 | 0.99 |
ENST00000479229.1
|
RP11-731F5.1
|
RP11-731F5.1 |
| chr20_+_44098346 | 0.96 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr6_-_46922659 | 0.95 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr13_-_36705425 | 0.94 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr19_+_35630022 | 0.91 |
ENST00000589209.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr9_-_117150243 | 0.90 |
ENST00000374088.3
|
AKNA
|
AT-hook transcription factor |
| chr3_-_49170405 | 0.89 |
ENST00000305544.4
ENST00000494831.1 |
LAMB2
|
laminin, beta 2 (laminin S) |
| chr17_-_1531635 | 0.85 |
ENST00000571650.1
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr19_+_35630344 | 0.84 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr2_-_99757977 | 0.84 |
ENST00000355053.4
|
TSGA10
|
testis specific, 10 |
| chr19_-_20844343 | 0.84 |
ENST00000595405.1
|
ZNF626
|
zinc finger protein 626 |
| chr3_-_49170522 | 0.79 |
ENST00000418109.1
|
LAMB2
|
laminin, beta 2 (laminin S) |
| chr3_-_122134882 | 0.78 |
ENST00000330689.4
|
WDR5B
|
WD repeat domain 5B |
| chr3_+_69915385 | 0.78 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr10_-_61469837 | 0.76 |
ENST00000395348.3
|
SLC16A9
|
solute carrier family 16, member 9 |
| chrX_+_133371077 | 0.72 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr10_+_91589261 | 0.70 |
ENST00000448963.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr17_-_10633291 | 0.68 |
ENST00000578345.1
ENST00000455996.2 |
TMEM220
|
transmembrane protein 220 |
| chr3_-_66551397 | 0.68 |
ENST00000383703.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr19_-_12251202 | 0.64 |
ENST00000334213.5
|
ZNF20
|
zinc finger protein 20 |
| chr3_-_66551351 | 0.62 |
ENST00000273261.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr4_-_25865159 | 0.62 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr1_-_226129083 | 0.61 |
ENST00000420304.2
|
LEFTY2
|
left-right determination factor 2 |
| chr20_+_44098385 | 0.61 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr1_+_110198689 | 0.61 |
ENST00000369836.4
|
GSTM4
|
glutathione S-transferase mu 4 |
| chr7_+_30960915 | 0.60 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr1_-_226129189 | 0.60 |
ENST00000366820.5
|
LEFTY2
|
left-right determination factor 2 |
| chr12_+_56473939 | 0.59 |
ENST00000450146.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr17_+_1627834 | 0.58 |
ENST00000419248.1
ENST00000418841.1 |
WDR81
|
WD repeat domain 81 |
| chr2_-_99757876 | 0.56 |
ENST00000539964.1
ENST00000393482.3 |
TSGA10
|
testis specific, 10 |
| chr19_+_8455200 | 0.56 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr14_-_61190754 | 0.56 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chrX_+_69674943 | 0.55 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr19_-_12405606 | 0.53 |
ENST00000356109.5
|
ZNF44
|
zinc finger protein 44 |
| chr11_+_17298297 | 0.52 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2 |
| chr7_+_63774321 | 0.52 |
ENST00000423484.2
|
ZNF736
|
zinc finger protein 736 |
| chr11_+_17298255 | 0.52 |
ENST00000531172.1
ENST00000533738.2 ENST00000323688.6 |
NUCB2
|
nucleobindin 2 |
| chr14_+_59104741 | 0.52 |
ENST00000395153.3
ENST00000335867.4 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr2_+_98330009 | 0.49 |
ENST00000264972.5
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa |
| chr15_+_74218787 | 0.48 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr3_-_52488048 | 0.48 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr3_-_52443799 | 0.48 |
ENST00000470173.1
ENST00000296288.5 |
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr19_-_12405689 | 0.47 |
ENST00000355684.5
|
ZNF44
|
zinc finger protein 44 |
| chr15_-_65715401 | 0.47 |
ENST00000352385.2
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr19_+_12075844 | 0.46 |
ENST00000592625.1
ENST00000586494.1 ENST00000343949.5 ENST00000545530.1 ENST00000358987.3 |
ZNF763
|
zinc finger protein 763 |
| chr17_+_57408994 | 0.46 |
ENST00000312655.4
|
YPEL2
|
yippee-like 2 (Drosophila) |
| chrX_+_24167746 | 0.46 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr14_-_61124977 | 0.46 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr6_+_149068464 | 0.46 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr17_-_27278304 | 0.45 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr15_-_82338460 | 0.44 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr11_-_77122928 | 0.43 |
ENST00000528203.1
ENST00000528592.1 ENST00000528633.1 ENST00000529248.1 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr17_-_39191107 | 0.43 |
ENST00000344363.5
|
KRTAP1-3
|
keratin associated protein 1-3 |
| chr5_-_131826457 | 0.43 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chrX_+_24167828 | 0.42 |
ENST00000379188.3
ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX
|
zinc finger protein, X-linked |
| chr12_-_55042140 | 0.41 |
ENST00000293371.6
ENST00000456047.2 |
DCD
|
dermcidin |
| chr19_+_41305627 | 0.40 |
ENST00000593525.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr1_+_9599540 | 0.40 |
ENST00000302692.6
|
SLC25A33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr14_+_96671016 | 0.39 |
ENST00000542454.2
ENST00000554311.1 ENST00000306005.3 ENST00000539359.1 ENST00000553811.1 |
BDKRB2
RP11-404P21.8
|
bradykinin receptor B2 Uncharacterized protein |
| chr19_+_41305612 | 0.39 |
ENST00000594380.1
ENST00000593397.1 ENST00000601733.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr19_+_8455077 | 0.38 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr15_-_62352570 | 0.38 |
ENST00000261517.5
ENST00000395896.4 ENST00000395898.3 |
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr12_+_56473628 | 0.38 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr19_+_41305330 | 0.38 |
ENST00000593972.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr12_+_82752275 | 0.38 |
ENST00000248306.3
|
METTL25
|
methyltransferase like 25 |
| chr6_-_32731299 | 0.37 |
ENST00000435145.2
ENST00000437316.2 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr1_+_204839959 | 0.37 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr20_-_48530230 | 0.37 |
ENST00000422556.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr3_+_61547585 | 0.37 |
ENST00000295874.10
ENST00000474889.1 |
PTPRG
|
protein tyrosine phosphatase, receptor type, G |
| chr12_+_56473910 | 0.37 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr17_-_42452063 | 0.36 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr19_-_55686399 | 0.36 |
ENST00000587067.1
|
SYT5
|
synaptotagmin V |
| chr6_-_30710265 | 0.36 |
ENST00000438162.1
ENST00000454845.1 |
FLOT1
|
flotillin 1 |
| chr7_-_112726393 | 0.35 |
ENST00000449591.1
ENST00000449735.1 ENST00000438062.1 ENST00000424100.1 |
GPR85
|
G protein-coupled receptor 85 |
| chr19_-_12662314 | 0.35 |
ENST00000339282.7
ENST00000596193.1 |
ZNF564
|
zinc finger protein 564 |
| chr4_-_2264015 | 0.34 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr11_+_1856034 | 0.34 |
ENST00000341958.3
|
SYT8
|
synaptotagmin VIII |
| chr21_-_31588365 | 0.34 |
ENST00000399899.1
|
CLDN8
|
claudin 8 |
| chr1_-_98386543 | 0.34 |
ENST00000423006.2
ENST00000370192.3 ENST00000306031.5 |
DPYD
|
dihydropyrimidine dehydrogenase |
| chr12_+_52695617 | 0.33 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr21_-_31588338 | 0.33 |
ENST00000286809.1
|
CLDN8
|
claudin 8 |
| chr10_+_123923205 | 0.32 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr8_+_49984894 | 0.32 |
ENST00000522267.1
ENST00000399653.4 ENST00000303202.8 |
C8orf22
|
chromosome 8 open reading frame 22 |
| chr22_-_38484922 | 0.32 |
ENST00000428572.1
|
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr17_+_38171681 | 0.32 |
ENST00000225474.2
ENST00000331769.2 ENST00000394148.3 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr12_-_66563831 | 0.31 |
ENST00000358230.3
|
TMBIM4
|
transmembrane BAX inhibitor motif containing 4 |
| chr19_+_41305406 | 0.31 |
ENST00000406058.2
ENST00000593726.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr5_+_98109322 | 0.31 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr19_+_23299777 | 0.31 |
ENST00000597761.2
|
ZNF730
|
zinc finger protein 730 |
| chr17_+_7792101 | 0.31 |
ENST00000358181.4
ENST00000330494.7 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr10_+_123922941 | 0.30 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr7_-_64023441 | 0.30 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr1_+_110198714 | 0.30 |
ENST00000336075.5
ENST00000326729.5 |
GSTM4
|
glutathione S-transferase mu 4 |
| chr15_+_74466744 | 0.30 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr15_+_60296421 | 0.30 |
ENST00000396057.4
|
FOXB1
|
forkhead box B1 |
| chr1_+_160121356 | 0.29 |
ENST00000368081.4
|
ATP1A4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr20_+_55204351 | 0.29 |
ENST00000201031.2
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr11_+_65265141 | 0.29 |
ENST00000534336.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr19_+_41305085 | 0.29 |
ENST00000303961.4
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr12_-_66563786 | 0.29 |
ENST00000542724.1
|
TMBIM4
|
transmembrane BAX inhibitor motif containing 4 |
| chr6_-_30709980 | 0.28 |
ENST00000416018.1
ENST00000445853.1 ENST00000413165.1 ENST00000418160.1 |
FLOT1
|
flotillin 1 |
| chr8_-_145060593 | 0.27 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr6_-_52860171 | 0.27 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr5_-_139422654 | 0.27 |
ENST00000289409.4
ENST00000358522.3 ENST00000378238.4 ENST00000289422.7 ENST00000361474.1 ENST00000545385.1 ENST00000394770.1 ENST00000541337.1 |
NRG2
|
neuregulin 2 |
| chr17_+_55183261 | 0.26 |
ENST00000576295.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr19_+_24269981 | 0.26 |
ENST00000339642.6
ENST00000357002.4 |
ZNF254
|
zinc finger protein 254 |
| chr11_+_67776012 | 0.26 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr19_-_4338783 | 0.25 |
ENST00000601482.1
ENST00000600324.1 |
STAP2
|
signal transducing adaptor family member 2 |
| chr16_+_22019404 | 0.25 |
ENST00000542527.2
ENST00000569656.1 ENST00000562695.1 |
C16orf52
|
chromosome 16 open reading frame 52 |
| chr9_+_108424738 | 0.25 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr5_+_145583156 | 0.25 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr19_+_10563567 | 0.25 |
ENST00000344979.3
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr8_-_27850141 | 0.24 |
ENST00000524352.1
|
SCARA5
|
scavenger receptor class A, member 5 (putative) |
| chr9_-_130829588 | 0.24 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr12_+_49687425 | 0.24 |
ENST00000257860.4
|
PRPH
|
peripherin |
| chr1_-_111743285 | 0.24 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr10_+_123923105 | 0.23 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr4_+_83351715 | 0.23 |
ENST00000273920.3
|
ENOPH1
|
enolase-phosphatase 1 |
| chr2_-_233415220 | 0.23 |
ENST00000408957.3
|
TIGD1
|
tigger transposable element derived 1 |
| chr7_-_43965937 | 0.23 |
ENST00000455877.1
ENST00000223341.7 ENST00000447717.3 ENST00000426198.1 |
URGCP
|
upregulator of cell proliferation |
| chr19_-_12662240 | 0.23 |
ENST00000416136.1
ENST00000428311.1 |
ZNF564
ZNF709
|
zinc finger protein 564 ZNF709 |
| chr1_+_166958497 | 0.22 |
ENST00000367870.2
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr19_-_4338838 | 0.22 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr5_+_79703823 | 0.22 |
ENST00000338008.5
ENST00000510158.1 ENST00000505560.1 |
ZFYVE16
|
zinc finger, FYVE domain containing 16 |
| chr16_+_333152 | 0.22 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr11_+_96123158 | 0.22 |
ENST00000332349.4
ENST00000458427.1 |
JRKL
|
jerky homolog-like (mouse) |
| chr17_-_33775760 | 0.22 |
ENST00000534689.1
ENST00000532210.1 ENST00000526861.1 ENST00000531588.1 ENST00000285013.6 |
SLFN13
|
schlafen family member 13 |
| chr2_+_111878483 | 0.21 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr19_-_49843539 | 0.21 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr16_+_30710462 | 0.21 |
ENST00000262518.4
ENST00000395059.2 ENST00000344771.4 |
SRCAP
|
Snf2-related CREBBP activator protein |
| chr19_+_12721725 | 0.21 |
ENST00000446165.1
ENST00000343325.4 ENST00000458122.3 |
ZNF791
|
zinc finger protein 791 |
| chr12_+_10365404 | 0.21 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr17_+_57642886 | 0.21 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr14_+_32546274 | 0.21 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr19_-_12595586 | 0.20 |
ENST00000397732.3
|
ZNF709
|
zinc finger protein 709 |
| chr1_-_154531095 | 0.20 |
ENST00000292211.4
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr22_+_40440804 | 0.20 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr19_-_39421377 | 0.20 |
ENST00000430193.3
ENST00000600042.1 ENST00000221431.6 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr12_-_48499591 | 0.20 |
ENST00000551330.1
ENST00000004980.5 ENST00000339976.6 ENST00000448372.1 |
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr19_+_13988061 | 0.19 |
ENST00000339133.5
ENST00000397555.2 |
NANOS3
|
nanos homolog 3 (Drosophila) |
| chrX_-_154688276 | 0.19 |
ENST00000369445.2
|
F8A3
|
coagulation factor VIII-associated 3 |
| chr3_-_48956818 | 0.19 |
ENST00000408959.2
|
ARIH2OS
|
ariadne homolog 2 opposite strand |
| chr7_-_122635754 | 0.19 |
ENST00000249284.2
|
TAS2R16
|
taste receptor, type 2, member 16 |
| chr22_+_50986462 | 0.19 |
ENST00000395676.2
|
KLHDC7B
|
kelch domain containing 7B |
| chr9_+_116111794 | 0.19 |
ENST00000374183.4
|
BSPRY
|
B-box and SPRY domain containing |
| chr14_+_64970662 | 0.19 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr6_-_52859968 | 0.18 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr1_-_35658736 | 0.18 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr2_-_20189819 | 0.18 |
ENST00000281405.4
ENST00000345530.3 |
WDR35
|
WD repeat domain 35 |
| chr17_-_60142609 | 0.18 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr14_-_106994333 | 0.18 |
ENST00000390624.2
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr16_-_69419473 | 0.18 |
ENST00000566750.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr5_+_145583107 | 0.18 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr2_-_27603582 | 0.18 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr1_+_55107449 | 0.17 |
ENST00000421030.2
ENST00000545244.1 ENST00000339553.5 ENST00000409996.1 ENST00000454855.2 |
MROH7
|
maestro heat-like repeat family member 7 |
| chr3_-_50605150 | 0.17 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr11_+_18344106 | 0.17 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr9_+_131451480 | 0.17 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chr5_+_177631523 | 0.17 |
ENST00000506339.1
ENST00000355836.5 ENST00000514633.1 ENST00000515193.1 ENST00000506259.1 ENST00000504898.1 |
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr22_+_25595817 | 0.17 |
ENST00000215855.2
ENST00000404334.1 |
CRYBB3
|
crystallin, beta B3 |
| chr5_-_169626104 | 0.17 |
ENST00000520275.1
ENST00000506431.2 |
CTB-27N1.1
|
CTB-27N1.1 |
| chr17_-_39183452 | 0.17 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr1_-_11107280 | 0.17 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr17_-_27278445 | 0.17 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr11_-_76155700 | 0.17 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr19_+_55795493 | 0.16 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr16_+_3508063 | 0.16 |
ENST00000576787.1
ENST00000572942.1 ENST00000576916.1 ENST00000575076.1 ENST00000572131.1 |
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr19_-_36297632 | 0.16 |
ENST00000588266.2
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr11_-_96123022 | 0.16 |
ENST00000542662.1
|
CCDC82
|
coiled-coil domain containing 82 |
| chr2_-_56150910 | 0.16 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chrX_-_99891796 | 0.16 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr1_-_28241226 | 0.15 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr11_-_128737259 | 0.15 |
ENST00000440599.2
ENST00000392666.1 ENST00000324036.3 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr5_+_177631497 | 0.15 |
ENST00000358344.3
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr12_+_57849048 | 0.15 |
ENST00000266646.2
|
INHBE
|
inhibin, beta E |
| chr11_-_128737163 | 0.15 |
ENST00000324003.3
ENST00000392665.2 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr8_-_103876965 | 0.14 |
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1 |
| chr3_-_50605077 | 0.14 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr1_-_89458636 | 0.14 |
ENST00000370486.1
ENST00000399794.2 |
CCBL2
RBMXL1
|
cysteine conjugate-beta lyase 2 RNA binding motif protein, X-linked-like 1 |
| chr1_-_43751230 | 0.14 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr1_-_161519579 | 0.14 |
ENST00000426740.1
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr17_+_42264395 | 0.14 |
ENST00000587989.1
ENST00000590235.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr1_-_89458415 | 0.14 |
ENST00000321792.5
ENST00000370491.3 |
RBMXL1
CCBL2
|
RNA binding motif protein, X-linked-like 1 cysteine conjugate-beta lyase 2 |
| chr17_+_16318909 | 0.14 |
ENST00000577397.1
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2 |
| chr4_-_76862117 | 0.14 |
ENST00000507956.1
ENST00000507187.2 ENST00000399497.3 ENST00000286733.4 |
NAAA
|
N-acylethanolamine acid amidase |
| chr10_+_35416223 | 0.14 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr11_+_65190245 | 0.14 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr10_-_98480243 | 0.13 |
ENST00000339364.5
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr19_+_11877838 | 0.13 |
ENST00000357901.4
ENST00000454339.2 |
ZNF441
|
zinc finger protein 441 |
| chr20_+_30458431 | 0.13 |
ENST00000375938.4
ENST00000535842.1 ENST00000310998.4 ENST00000375921.2 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr17_+_62503147 | 0.13 |
ENST00000553412.1
|
CEP95
|
centrosomal protein 95kDa |
| chr17_+_42264322 | 0.13 |
ENST00000446571.3
ENST00000357984.3 ENST00000538716.2 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr8_-_124553437 | 0.13 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2L1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.3 | 1.7 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.3 | 1.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.6 | GO:0072229 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.2 | 0.6 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 | 4.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 0.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.4 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.4 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.5 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.5 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.5 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.4 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 | 0.9 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.9 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 0.5 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.3 | GO:0019860 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.5 | GO:0043366 | beta selection(GO:0043366) |
| 0.1 | 0.4 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 0.2 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.3 | GO:0010847 | regulation of chromatin assembly(GO:0010847) negative regulation of protein import into nucleus, translocation(GO:0033159) protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.2 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.2 | GO:0032213 | regulation of telomere maintenance via semi-conservative replication(GO:0032213) negative regulation of telomere maintenance via semi-conservative replication(GO:0032214) |
| 0.1 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 1.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.8 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.3 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 1.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 1.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.3 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.3 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.4 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.5 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.4 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.4 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0002433 | Fc receptor mediated stimulatory signaling pathway(GO:0002431) immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.2 | 0.6 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 3.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.2 | GO:0030849 | autosome(GO:0030849) |
| 0.1 | 0.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 1.3 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.2 | 0.6 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 1.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 1.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.9 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 2.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 1.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 1.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 3.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.9 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |