Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for TP63
Z-value: 1.34
Transcription factors associated with TP63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP63
|
ENSG00000073282.8 | tumor protein p63 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP63 | hg19_v2_chr3_+_189349162_189349305 | 0.76 | 1.0e-06 | Click! |
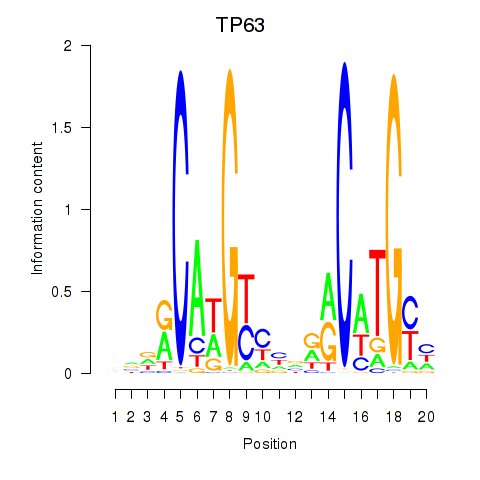
Activity profile of TP63 motif
Sorted Z-values of TP63 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51504411 | 14.45 |
ENST00000593490.1
|
KLK8
|
kallikrein-related peptidase 8 |
| chr19_-_51471381 | 8.57 |
ENST00000594641.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr19_-_19051103 | 5.60 |
ENST00000542541.2
ENST00000433218.2 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr2_-_216003127 | 4.94 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr1_+_151030234 | 4.78 |
ENST00000368921.3
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11 |
| chr12_-_52845910 | 4.54 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr20_-_43280325 | 4.51 |
ENST00000537820.1
|
ADA
|
adenosine deaminase |
| chr4_-_10023095 | 4.43 |
ENST00000264784.3
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr12_+_119616447 | 4.17 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr2_-_31361543 | 4.00 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr3_-_50340996 | 3.96 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr1_+_17575584 | 3.88 |
ENST00000375460.3
|
PADI3
|
peptidyl arginine deiminase, type III |
| chr1_-_26197744 | 3.79 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII |
| chr20_-_43280361 | 3.63 |
ENST00000372874.4
|
ADA
|
adenosine deaminase |
| chr10_+_102106829 | 3.61 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chrX_-_153602991 | 3.50 |
ENST00000369850.3
ENST00000422373.1 |
FLNA
|
filamin A, alpha |
| chr1_+_86889769 | 3.43 |
ENST00000370565.4
|
CLCA2
|
chloride channel accessory 2 |
| chr18_+_34124507 | 3.42 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr1_-_47655686 | 3.33 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr1_-_228603694 | 3.27 |
ENST00000366697.2
|
TRIM17
|
tripartite motif containing 17 |
| chr19_+_18496957 | 3.17 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr22_+_31489344 | 3.05 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr7_+_80267973 | 2.95 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr20_-_52790512 | 2.89 |
ENST00000216862.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr10_+_99344104 | 2.73 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr18_+_61143994 | 2.49 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr20_-_6103666 | 2.47 |
ENST00000536936.1
|
FERMT1
|
fermitin family member 1 |
| chr19_-_11688447 | 2.43 |
ENST00000590420.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr22_+_38302285 | 2.38 |
ENST00000215957.6
|
MICALL1
|
MICAL-like 1 |
| chr12_-_125348329 | 2.35 |
ENST00000546215.1
ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1
|
scavenger receptor class B, member 1 |
| chr1_+_38158090 | 2.34 |
ENST00000373055.1
ENST00000327331.2 |
CDCA8
|
cell division cycle associated 8 |
| chr6_+_106546808 | 2.27 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr10_-_90751038 | 2.24 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr17_+_26662597 | 2.23 |
ENST00000544907.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr6_+_121756809 | 2.20 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chrX_+_2670066 | 2.19 |
ENST00000381174.5
ENST00000419513.2 ENST00000426774.1 |
XG
|
Xg blood group |
| chr12_+_120105558 | 2.16 |
ENST00000229328.5
ENST00000541640.1 |
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr8_-_23021533 | 2.12 |
ENST00000312584.3
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
| chr19_-_11688500 | 2.04 |
ENST00000433365.2
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr8_+_22960426 | 2.00 |
ENST00000540813.1
|
TNFRSF10C
|
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain |
| chr19_+_535835 | 1.97 |
ENST00000607527.1
ENST00000606065.1 |
CDC34
|
cell division cycle 34 |
| chr7_+_102389434 | 1.96 |
ENST00000409231.3
ENST00000418198.1 |
FAM185A
|
family with sequence similarity 185, member A |
| chr17_+_26662730 | 1.96 |
ENST00000226225.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chrX_+_150148976 | 1.96 |
ENST00000419110.1
|
HMGB3
|
high mobility group box 3 |
| chr12_-_125348448 | 1.95 |
ENST00000339570.5
|
SCARB1
|
scavenger receptor class B, member 1 |
| chr8_-_22926526 | 1.94 |
ENST00000347739.3
ENST00000542226.1 |
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b |
| chr1_+_24645865 | 1.90 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_24645807 | 1.87 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_24646002 | 1.85 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr14_+_51955831 | 1.80 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
| chr9_+_35906176 | 1.78 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr6_-_30712313 | 1.78 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr11_-_111782484 | 1.77 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr15_-_89456630 | 1.74 |
ENST00000268150.8
|
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chr11_+_77899920 | 1.71 |
ENST00000528910.1
ENST00000529308.1 |
USP35
|
ubiquitin specific peptidase 35 |
| chr10_-_105212059 | 1.69 |
ENST00000260743.5
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr2_-_165698662 | 1.69 |
ENST00000194871.6
ENST00000445474.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chrX_+_152086373 | 1.63 |
ENST00000318529.8
|
ZNF185
|
zinc finger protein 185 (LIM domain) |
| chr11_+_65686728 | 1.61 |
ENST00000312515.2
ENST00000525501.1 |
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr19_+_48824711 | 1.61 |
ENST00000599704.1
|
EMP3
|
epithelial membrane protein 3 |
| chr2_+_234600253 | 1.58 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr14_-_94595993 | 1.58 |
ENST00000238609.3
|
IFI27L2
|
interferon, alpha-inducible protein 27-like 2 |
| chr17_+_75450075 | 1.55 |
ENST00000592951.1
|
SEPT9
|
septin 9 |
| chr15_-_89456593 | 1.50 |
ENST00000558029.1
ENST00000539437.1 ENST00000542878.1 ENST00000268151.7 ENST00000566497.1 |
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chr17_+_80332153 | 1.49 |
ENST00000313135.2
|
UTS2R
|
urotensin 2 receptor |
| chr22_+_24999114 | 1.48 |
ENST00000412658.1
ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr8_-_49833978 | 1.47 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr12_-_96184533 | 1.47 |
ENST00000343702.4
ENST00000344911.4 |
NTN4
|
netrin 4 |
| chr10_-_105212141 | 1.43 |
ENST00000369788.3
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr1_-_19229248 | 1.43 |
ENST00000375341.3
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr4_-_21546272 | 1.42 |
ENST00000509207.1
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr1_-_93426998 | 1.42 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr1_-_19229014 | 1.39 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr1_-_153588334 | 1.29 |
ENST00000476873.1
|
S100A14
|
S100 calcium binding protein A14 |
| chr9_+_116298778 | 1.28 |
ENST00000462143.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chr3_+_136676851 | 1.27 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr9_+_139553306 | 1.26 |
ENST00000371699.1
|
EGFL7
|
EGF-like-domain, multiple 7 |
| chr8_-_22926623 | 1.25 |
ENST00000276431.4
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b |
| chr3_-_45017609 | 1.25 |
ENST00000342790.4
ENST00000424952.2 ENST00000296127.3 ENST00000455235.1 |
ZDHHC3
|
zinc finger, DHHC-type containing 3 |
| chr1_-_209824643 | 1.23 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr15_-_42448788 | 1.21 |
ENST00000382396.4
ENST00000397272.3 |
PLA2G4F
|
phospholipase A2, group IVF |
| chr3_+_136676707 | 1.17 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr1_-_152552980 | 1.15 |
ENST00000368787.3
|
LCE3D
|
late cornified envelope 3D |
| chr16_+_23652773 | 1.15 |
ENST00000563998.1
ENST00000568589.1 ENST00000568272.1 |
DCTN5
|
dynactin 5 (p25) |
| chr3_+_44379611 | 1.12 |
ENST00000383746.3
ENST00000417237.1 |
TCAIM
|
T cell activation inhibitor, mitochondrial |
| chr4_+_3388057 | 1.11 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr12_-_52887034 | 1.11 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr2_-_165698521 | 1.10 |
ENST00000409184.3
ENST00000392717.2 ENST00000456693.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr2_-_47168850 | 1.09 |
ENST00000409207.1
|
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr11_+_18287801 | 1.08 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr11_+_18287721 | 1.07 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr1_-_94312706 | 1.07 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr5_+_153570319 | 1.06 |
ENST00000377661.2
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr16_+_8807419 | 1.06 |
ENST00000565016.1
ENST00000567812.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr4_+_2794750 | 1.02 |
ENST00000452765.2
ENST00000389838.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr4_+_8321882 | 1.02 |
ENST00000509453.1
ENST00000503186.1 |
RP11-774O3.2
RP11-774O3.1
|
RP11-774O3.2 RP11-774O3.1 |
| chr8_+_22853345 | 1.01 |
ENST00000522948.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr19_+_49458107 | 1.00 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr3_-_33700589 | 1.00 |
ENST00000461133.3
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr6_+_54711533 | 1.00 |
ENST00000306858.7
|
FAM83B
|
family with sequence similarity 83, member B |
| chr13_+_42846272 | 0.98 |
ENST00000025301.2
|
AKAP11
|
A kinase (PRKA) anchor protein 11 |
| chr20_+_30467600 | 0.97 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr6_+_134758827 | 0.96 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr16_+_1832902 | 0.96 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr15_+_68570062 | 0.95 |
ENST00000306917.4
|
FEM1B
|
fem-1 homolog b (C. elegans) |
| chr7_+_23145884 | 0.94 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr21_+_44313375 | 0.94 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr19_+_35861831 | 0.92 |
ENST00000454971.1
|
GPR42
|
G protein-coupled receptor 42 (gene/pseudogene) |
| chr19_+_35862192 | 0.92 |
ENST00000597214.1
|
GPR42
|
G protein-coupled receptor 42 (gene/pseudogene) |
| chr1_+_154300217 | 0.90 |
ENST00000368489.3
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr6_-_44923160 | 0.89 |
ENST00000371458.1
|
SUPT3H
|
suppressor of Ty 3 homolog (S. cerevisiae) |
| chr19_+_35849362 | 0.88 |
ENST00000327809.4
|
FFAR3
|
free fatty acid receptor 3 |
| chr11_+_61891445 | 0.87 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr6_-_101329157 | 0.84 |
ENST00000369143.2
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr15_-_55700216 | 0.84 |
ENST00000569205.1
|
CCPG1
|
cell cycle progression 1 |
| chr8_-_117778494 | 0.84 |
ENST00000276682.4
|
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr2_+_131369054 | 0.84 |
ENST00000409602.1
|
POTEJ
|
POTE ankyrin domain family, member J |
| chr3_-_57113314 | 0.83 |
ENST00000338458.4
ENST00000468727.1 |
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr2_+_47168630 | 0.83 |
ENST00000263737.6
|
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr19_+_42364460 | 0.82 |
ENST00000593863.1
|
RPS19
|
ribosomal protein S19 |
| chr20_+_57264187 | 0.82 |
ENST00000525967.1
ENST00000525817.1 |
NPEPL1
|
aminopeptidase-like 1 |
| chr16_+_30007524 | 0.81 |
ENST00000567254.1
ENST00000567705.1 |
INO80E
|
INO80 complex subunit E |
| chr16_+_30006997 | 0.81 |
ENST00000304516.7
|
INO80E
|
INO80 complex subunit E |
| chr19_-_46288917 | 0.81 |
ENST00000537879.1
ENST00000596586.1 ENST00000595946.1 |
DMWD
AC011530.4
|
dystrophia myotonica, WD repeat containing Uncharacterized protein |
| chr3_+_184080387 | 0.80 |
ENST00000455712.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr22_+_50628999 | 0.80 |
ENST00000395827.1
|
TRABD
|
TraB domain containing |
| chr5_+_153570285 | 0.79 |
ENST00000425427.2
ENST00000297107.6 |
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr19_+_35849723 | 0.79 |
ENST00000594310.1
|
FFAR3
|
free fatty acid receptor 3 |
| chr11_-_111782696 | 0.77 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr18_+_21033239 | 0.77 |
ENST00000581585.1
ENST00000577501.1 |
RIOK3
|
RIO kinase 3 |
| chr14_-_37642016 | 0.76 |
ENST00000331299.5
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr11_-_8190534 | 0.75 |
ENST00000309737.6
ENST00000425599.2 ENST00000539720.1 ENST00000531450.1 ENST00000419822.2 ENST00000335425.7 ENST00000343202.4 |
RIC3
|
RIC3 acetylcholine receptor chaperone |
| chr10_+_75532028 | 0.73 |
ENST00000372841.3
ENST00000394790.1 |
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
| chr6_-_101329191 | 0.72 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr11_+_18433840 | 0.72 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr10_+_90750493 | 0.72 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr19_+_42364313 | 0.71 |
ENST00000601492.1
ENST00000600467.1 ENST00000221975.2 |
RPS19
|
ribosomal protein S19 |
| chr2_-_130878140 | 0.70 |
ENST00000360967.5
ENST00000361163.4 ENST00000357462.5 |
POTEF
|
POTE ankyrin domain family, member F |
| chr21_-_27462351 | 0.70 |
ENST00000448850.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr12_+_69202975 | 0.70 |
ENST00000544561.1
ENST00000393410.1 ENST00000299252.4 ENST00000360430.2 ENST00000517852.1 ENST00000545204.1 ENST00000393413.3 ENST00000350057.5 ENST00000348801.2 ENST00000478070.1 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr17_-_74707037 | 0.70 |
ENST00000355797.3
ENST00000375036.2 ENST00000449428.2 |
MXRA7
|
matrix-remodelling associated 7 |
| chr7_-_72742085 | 0.69 |
ENST00000333149.2
|
TRIM50
|
tripartite motif containing 50 |
| chr10_+_11059826 | 0.69 |
ENST00000450189.1
|
CELF2
|
CUGBP, Elav-like family member 2 |
| chr12_-_126467906 | 0.68 |
ENST00000507313.1
ENST00000545784.1 |
LINC00939
|
long intergenic non-protein coding RNA 939 |
| chr15_+_32907691 | 0.68 |
ENST00000361627.3
ENST00000567348.1 ENST00000563864.1 ENST00000543522.1 |
ARHGAP11A
|
Rho GTPase activating protein 11A |
| chr2_-_47168906 | 0.66 |
ENST00000444761.2
ENST00000409147.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr8_+_126442563 | 0.66 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr12_+_69202795 | 0.66 |
ENST00000539479.1
ENST00000393415.3 ENST00000523991.1 ENST00000543323.1 ENST00000393416.2 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr16_-_68000717 | 0.63 |
ENST00000541864.2
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr17_+_79859985 | 0.62 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chr3_+_45017722 | 0.60 |
ENST00000265564.7
|
EXOSC7
|
exosome component 7 |
| chr16_-_47493041 | 0.59 |
ENST00000565940.2
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr19_+_19779686 | 0.59 |
ENST00000415784.2
|
ZNF101
|
zinc finger protein 101 |
| chr2_+_108994466 | 0.59 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr2_-_112614424 | 0.58 |
ENST00000427997.1
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr5_+_170814803 | 0.58 |
ENST00000521672.1
ENST00000351986.6 ENST00000393820.2 ENST00000523622.1 |
NPM1
|
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
| chr16_+_30006615 | 0.58 |
ENST00000563197.1
|
INO80E
|
INO80 complex subunit E |
| chr20_+_35234137 | 0.58 |
ENST00000344795.3
ENST00000373852.5 |
C20orf24
|
chromosome 20 open reading frame 24 |
| chr17_-_14140166 | 0.57 |
ENST00000420162.2
ENST00000431716.2 |
CDRT15
|
CMT1A duplicated region transcript 15 |
| chr7_+_23145366 | 0.57 |
ENST00000339077.5
ENST00000322275.5 ENST00000539124.1 ENST00000542558.1 |
KLHL7
|
kelch-like family member 7 |
| chr14_+_24779376 | 0.57 |
ENST00000530080.1
|
LTB4R2
|
leukotriene B4 receptor 2 |
| chr22_-_38577782 | 0.56 |
ENST00000430886.1
ENST00000332509.3 ENST00000447598.2 ENST00000435484.1 ENST00000402064.1 ENST00000436218.1 |
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr16_-_34740829 | 0.56 |
ENST00000562591.1
|
RP11-80F22.15
|
RP11-80F22.15 |
| chr7_-_142207004 | 0.56 |
ENST00000426318.2
|
TRBV10-2
|
T cell receptor beta variable 10-2 |
| chr20_+_35234223 | 0.56 |
ENST00000342422.3
|
C20orf24
|
chromosome 20 open reading frame 24 |
| chr3_+_47844399 | 0.55 |
ENST00000446256.2
ENST00000445061.1 |
DHX30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr10_+_124913793 | 0.54 |
ENST00000368865.4
ENST00000538238.1 ENST00000368859.2 |
BUB3
|
BUB3 mitotic checkpoint protein |
| chr6_+_31691121 | 0.53 |
ENST00000480039.1
ENST00000375810.4 ENST00000375805.2 ENST00000375809.3 ENST00000375804.2 ENST00000375814.3 ENST00000375806.2 |
C6orf25
|
chromosome 6 open reading frame 25 |
| chr9_+_93589734 | 0.53 |
ENST00000375746.1
|
SYK
|
spleen tyrosine kinase |
| chr4_-_144826682 | 0.53 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr7_+_72742178 | 0.52 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr1_+_12040238 | 0.52 |
ENST00000444836.1
ENST00000235329.5 |
MFN2
|
mitofusin 2 |
| chr17_+_49230897 | 0.52 |
ENST00000393196.3
ENST00000336097.3 ENST00000480143.1 ENST00000511355.1 ENST00000013034.3 ENST00000393198.3 ENST00000608447.1 ENST00000393193.2 ENST00000376392.6 ENST00000555572.1 |
NME1
NME1-NME2
NME2
|
NME/NM23 nucleoside diphosphate kinase 1 NME1-NME2 readthrough NME/NM23 nucleoside diphosphate kinase 2 |
| chr10_+_103113840 | 0.51 |
ENST00000393441.4
ENST00000408038.2 |
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr16_+_23652700 | 0.51 |
ENST00000300087.2
|
DCTN5
|
dynactin 5 (p25) |
| chr16_+_28834531 | 0.51 |
ENST00000570200.1
|
ATXN2L
|
ataxin 2-like |
| chr3_+_19189946 | 0.51 |
ENST00000328405.2
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr9_-_136605042 | 0.50 |
ENST00000371872.4
ENST00000298628.5 ENST00000422262.2 |
SARDH
|
sarcosine dehydrogenase |
| chrX_+_48334549 | 0.50 |
ENST00000019019.2
ENST00000348411.2 ENST00000396894.4 |
FTSJ1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr16_+_58010339 | 0.49 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr9_-_35619539 | 0.49 |
ENST00000396757.1
|
CD72
|
CD72 molecule |
| chr22_-_50946113 | 0.49 |
ENST00000216080.5
ENST00000474879.2 ENST00000380796.3 |
LMF2
|
lipase maturation factor 2 |
| chr4_+_15376165 | 0.48 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr1_-_20987982 | 0.48 |
ENST00000375048.3
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr21_+_43823983 | 0.48 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr11_-_1330834 | 0.47 |
ENST00000525159.1
ENST00000317204.6 ENST00000542915.1 ENST00000527938.1 ENST00000530541.1 ENST00000263646.7 |
TOLLIP
|
toll interacting protein |
| chr10_+_103113802 | 0.47 |
ENST00000370187.3
|
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chrX_-_129244454 | 0.47 |
ENST00000308167.5
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr1_-_114447620 | 0.47 |
ENST00000369569.1
ENST00000432415.1 ENST00000369571.2 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr19_-_10420459 | 0.46 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr17_-_79818354 | 0.46 |
ENST00000576541.1
ENST00000576380.1 ENST00000571617.1 ENST00000576052.1 ENST00000576390.1 ENST00000573778.2 ENST00000439918.2 ENST00000574914.1 ENST00000331483.4 |
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr17_+_21279509 | 0.45 |
ENST00000583088.1
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr7_+_72742162 | 0.45 |
ENST00000431982.2
|
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr5_-_179051579 | 0.45 |
ENST00000505811.1
ENST00000515714.1 ENST00000513225.1 ENST00000503664.1 ENST00000356731.5 ENST00000523137.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr22_-_38577731 | 0.45 |
ENST00000335539.3
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr20_+_816695 | 0.43 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chrX_-_153637612 | 0.43 |
ENST00000369807.1
ENST00000369808.3 |
DNASE1L1
|
deoxyribonuclease I-like 1 |
| chr1_-_114447683 | 0.43 |
ENST00000256658.4
ENST00000369564.1 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr5_-_64920115 | 0.43 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr3_-_9005118 | 0.42 |
ENST00000264926.2
|
RAD18
|
RAD18 homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP63
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.1 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 1.8 | 14.4 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 1.4 | 4.3 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 1.2 | 4.9 | GO:0035627 | ceramide transport(GO:0035627) |
| 1.2 | 3.6 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.0 | 4.0 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.0 | 2.9 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.9 | 2.7 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.9 | 4.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.8 | 3.9 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.7 | 2.2 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.6 | 2.4 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.6 | 3.5 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.5 | 1.5 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.5 | 1.5 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.5 | 2.5 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.5 | 1.5 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.5 | 0.5 | GO:0051604 | protein maturation(GO:0051604) |
| 0.5 | 2.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.5 | 2.3 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.5 | 1.4 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.4 | 2.9 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.4 | 1.7 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.4 | 8.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.4 | 6.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 2.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.3 | 1.0 | GO:1903521 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) positive regulation of apoptotic DNA fragmentation(GO:1902512) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.3 | 5.6 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 1.5 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.3 | 1.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 1.1 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.3 | 1.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.3 | 1.8 | GO:0003383 | apical constriction(GO:0003383) |
| 0.3 | 4.8 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.2 | 4.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 1.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 5.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 0.7 | GO:0045658 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 | 1.0 | GO:0038127 | epidermal growth factor receptor signaling pathway(GO:0007173) ERBB signaling pathway(GO:0038127) |
| 0.2 | 0.2 | GO:1903384 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.2 | 0.8 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 2.0 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.2 | 1.5 | GO:0090237 | regulation of arachidonic acid secretion(GO:0090237) |
| 0.2 | 3.9 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.2 | 0.5 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.1 | 0.6 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 1.5 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 2.2 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 2.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.7 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 4.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.9 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.4 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 4.1 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.8 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 3.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.6 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.7 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.1 | 2.9 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 1.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.9 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 0.6 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.3 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.1 | 0.4 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.7 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.3 | GO:0060435 | branchiomeric skeletal muscle development(GO:0014707) bronchiole development(GO:0060435) |
| 0.1 | 0.5 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.1 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.4 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 0.1 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 0.6 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 1.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.8 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.1 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.1 | 2.9 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 0.6 | GO:0060699 | regulation of endoribonuclease activity(GO:0060699) |
| 0.1 | 0.2 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.1 | 5.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 1.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 1.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.4 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 1.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.1 | 0.1 | GO:1902903 | regulation of fibril organization(GO:1902903) |
| 0.1 | 0.3 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 3.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.2 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.1 | 0.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.2 | GO:0044828 | negative regulation of CREB transcription factor activity(GO:0032792) negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.8 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.2 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 1.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.4 | GO:0018095 | sperm axoneme assembly(GO:0007288) protein polyglutamylation(GO:0018095) |
| 0.0 | 0.5 | GO:0018202 | UTP biosynthetic process(GO:0006228) peptidyl-histidine modification(GO:0018202) |
| 0.0 | 1.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 1.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 2.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 2.2 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.4 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 1.0 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.4 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 1.9 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 3.7 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 4.0 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 1.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 1.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 1.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 1.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.7 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.9 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.3 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 3.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.7 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 3.0 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 1.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.8 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 2.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 4.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.4 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.0 | 1.4 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 1.1 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.0 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.3 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0031523 | Myb complex(GO:0031523) |
| 0.7 | 4.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.6 | 2.9 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.4 | 2.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 2.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 1.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 8.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 0.7 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.2 | 0.8 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 1.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 4.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 2.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 2.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.3 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.6 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 10.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 2.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 3.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.9 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.9 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.3 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 2.1 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 1.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 3.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 5.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.6 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 2.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 4.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 2.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 0.0 | 2.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 1.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 4.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 31.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.0 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.7 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 1.2 | 4.9 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 1.2 | 7.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 1.0 | 4.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.9 | 3.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.8 | 3.9 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.7 | 3.6 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.7 | 2.7 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.6 | 2.9 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.5 | 2.9 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.5 | 8.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.5 | 1.4 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.4 | 2.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.4 | 5.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.3 | 1.0 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.3 | 4.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 4.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 2.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.3 | 1.1 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.2 | 5.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 3.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 4.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 1.6 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.2 | 0.5 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.2 | 0.7 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.2 | 1.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 0.5 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.7 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.4 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 2.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 3.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.8 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.0 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.1 | 1.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 2.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 2.8 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 1.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 2.4 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 0.7 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.9 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 19.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 2.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 2.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.2 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.1 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.2 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.0 | 3.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 7.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 2.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 1.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 2.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.7 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 3.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 1.3 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 3.0 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 3.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.8 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 1.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 3.4 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 23.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 9.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 7.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 2.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 2.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 4.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 4.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 4.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 4.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 3.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 2.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 5.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 1.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 4.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 2.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 2.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 3.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 3.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 2.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 1.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 2.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |