Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
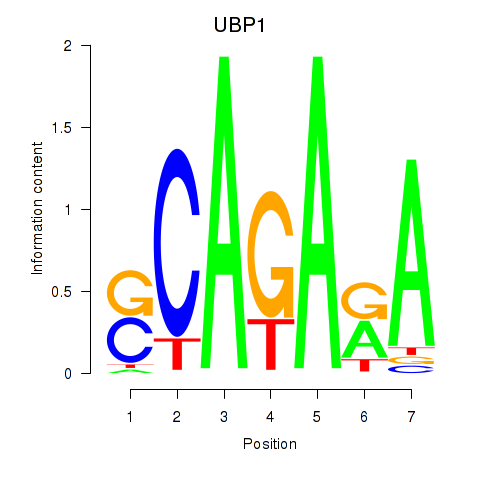
Results for UBP1
Z-value: 0.42
Transcription factors associated with UBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
UBP1
|
ENSG00000153560.7 | upstream binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| UBP1 | hg19_v2_chr3_-_33481835_33481905 | -0.60 | 4.1e-04 | Click! |
Activity profile of UBP1 motif
Sorted Z-values of UBP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_38071615 | 2.51 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr4_-_57524061 | 1.60 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr12_-_47473425 | 1.49 |
ENST00000550413.1
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr19_-_51538148 | 1.44 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chr17_+_74381343 | 1.36 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr2_+_228678550 | 1.30 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr19_+_917287 | 1.17 |
ENST00000592648.1
ENST00000234371.5 |
KISS1R
|
KISS1 receptor |
| chr17_-_5138099 | 0.94 |
ENST00000571800.1
ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr12_-_8815215 | 0.93 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr12_-_8815299 | 0.91 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr1_-_150208320 | 0.91 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr11_-_102668879 | 0.82 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr9_-_35685452 | 0.79 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 (beta) |
| chr12_+_100041527 | 0.69 |
ENST00000324341.1
|
FAM71C
|
family with sequence similarity 71, member C |
| chr12_+_47473369 | 0.67 |
ENST00000546455.1
|
PCED1B
|
PC-esterase domain containing 1B |
| chr19_+_41257084 | 0.65 |
ENST00000601393.1
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr2_-_89442621 | 0.57 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20 |
| chr10_-_75676400 | 0.54 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr19_-_49864746 | 0.53 |
ENST00000598810.1
|
TEAD2
|
TEA domain family member 2 |
| chrX_-_65253506 | 0.48 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr1_-_150208291 | 0.48 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr5_+_95998070 | 0.46 |
ENST00000421689.2
ENST00000510756.1 ENST00000512620.1 |
CAST
|
calpastatin |
| chr1_-_150208363 | 0.45 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr13_-_23949671 | 0.44 |
ENST00000402364.1
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr3_+_111718173 | 0.42 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr21_+_43823983 | 0.41 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr2_+_17721920 | 0.38 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr4_+_156587979 | 0.37 |
ENST00000511507.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr15_-_101835110 | 0.34 |
ENST00000560496.1
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr17_+_48450575 | 0.34 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr11_-_11374904 | 0.33 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr3_+_111718036 | 0.33 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr16_+_29823427 | 0.27 |
ENST00000358758.7
ENST00000567659.1 ENST00000572820.1 |
PRRT2
|
proline-rich transmembrane protein 2 |
| chr6_+_29429217 | 0.27 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr1_-_150208412 | 0.27 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_-_47009996 | 0.26 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr2_-_2334888 | 0.26 |
ENST00000428368.2
ENST00000399161.2 |
MYT1L
|
myelin transcription factor 1-like |
| chr6_+_31555045 | 0.26 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr4_+_156588115 | 0.26 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr11_-_60623437 | 0.24 |
ENST00000332539.4
|
PTGDR2
|
prostaglandin D2 receptor 2 |
| chr5_+_150051149 | 0.24 |
ENST00000523553.1
|
MYOZ3
|
myozenin 3 |
| chrX_+_38420623 | 0.23 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr6_-_29395509 | 0.23 |
ENST00000377147.2
|
OR11A1
|
olfactory receptor, family 11, subfamily A, member 1 |
| chr2_+_61244697 | 0.23 |
ENST00000401576.1
ENST00000295030.5 ENST00000414712.2 |
PEX13
|
peroxisomal biogenesis factor 13 |
| chr4_+_156587853 | 0.22 |
ENST00000506455.1
ENST00000511108.1 |
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chrX_+_78426469 | 0.21 |
ENST00000276077.1
|
GPR174
|
G protein-coupled receptor 174 |
| chr12_+_123717967 | 0.21 |
ENST00000536130.1
ENST00000546132.1 |
C12orf65
|
chromosome 12 open reading frame 65 |
| chr5_+_34656331 | 0.20 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr1_+_206223941 | 0.20 |
ENST00000367126.4
|
AVPR1B
|
arginine vasopressin receptor 1B |
| chr19_-_11039188 | 0.19 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr17_-_48450534 | 0.18 |
ENST00000503633.1
ENST00000442592.3 ENST00000225969.4 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr1_-_150208498 | 0.17 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr14_-_75330537 | 0.16 |
ENST00000556084.2
ENST00000556489.2 ENST00000445876.1 |
PROX2
|
prospero homeobox 2 |
| chr15_+_81475047 | 0.16 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr8_-_66474884 | 0.14 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr9_+_15422702 | 0.14 |
ENST00000380821.3
ENST00000421710.1 |
SNAPC3
|
small nuclear RNA activating complex, polypeptide 3, 50kDa |
| chr8_-_107782463 | 0.14 |
ENST00000311955.3
|
ABRA
|
actin-binding Rho activating protein |
| chr1_+_155108294 | 0.13 |
ENST00000303343.8
ENST00000368404.4 ENST00000368401.5 |
SLC50A1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chrX_+_139791917 | 0.13 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr1_-_153599732 | 0.12 |
ENST00000392623.1
|
S100A13
|
S100 calcium binding protein A13 |
| chr14_+_21214039 | 0.12 |
ENST00000326842.2
|
EDDM3A
|
epididymal protein 3A |
| chr1_-_156217875 | 0.12 |
ENST00000292291.5
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr15_+_28624878 | 0.11 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr3_+_111260980 | 0.11 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr1_-_182641367 | 0.10 |
ENST00000508450.1
|
RGS8
|
regulator of G-protein signaling 8 |
| chr12_+_123717458 | 0.10 |
ENST00000253233.1
|
C12orf65
|
chromosome 12 open reading frame 65 |
| chr8_+_77593448 | 0.10 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr19_-_54784937 | 0.10 |
ENST00000434421.1
ENST00000314446.5 ENST00000391749.4 |
LILRB2
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
| chr11_+_10326612 | 0.10 |
ENST00000534464.1
ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM
|
adrenomedullin |
| chr2_+_113033164 | 0.10 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr7_+_132937820 | 0.09 |
ENST00000393161.2
ENST00000253861.4 |
EXOC4
|
exocyst complex component 4 |
| chr3_+_111260954 | 0.08 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr11_-_113345995 | 0.08 |
ENST00000355319.2
ENST00000542616.1 |
DRD2
|
dopamine receptor D2 |
| chr19_-_51875523 | 0.08 |
ENST00000593572.1
ENST00000595157.1 |
NKG7
|
natural killer cell group 7 sequence |
| chr22_+_22676808 | 0.07 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr22_+_22730353 | 0.07 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr20_+_59654146 | 0.07 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr7_+_29874341 | 0.07 |
ENST00000409290.1
ENST00000242140.5 |
WIPF3
|
WAS/WASL interacting protein family, member 3 |
| chr1_+_48688357 | 0.06 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr2_+_162087577 | 0.06 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr5_-_41510656 | 0.05 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr1_-_152332480 | 0.05 |
ENST00000388718.5
|
FLG2
|
filaggrin family member 2 |
| chr3_+_111260856 | 0.05 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr19_-_11039261 | 0.04 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr10_-_27443294 | 0.04 |
ENST00000396296.3
ENST00000375972.3 ENST00000376016.3 ENST00000491542.2 |
YME1L1
|
YME1-like 1 ATPase |
| chr5_+_122847908 | 0.00 |
ENST00000511130.2
ENST00000512718.3 |
CSNK1G3
|
casein kinase 1, gamma 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of UBP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.5 | 1.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.4 | 1.3 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.8 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.1 | 0.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.2 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.2 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 1.6 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 1.8 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 2.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.5 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.7 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.5 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 1.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.8 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.1 | GO:1900168 | glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.6 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.3 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.1 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.4 | GO:0070268 | cornification(GO:0070268) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071749 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.1 | 0.5 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 2.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.7 | GO:0005685 | U1 snRNP(GO:0005685) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0030395 | lactose binding(GO:0030395) |
| 0.4 | 1.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 1.4 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.7 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 2.3 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 1.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.1 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 2.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |