Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for VDR
Z-value: 0.87
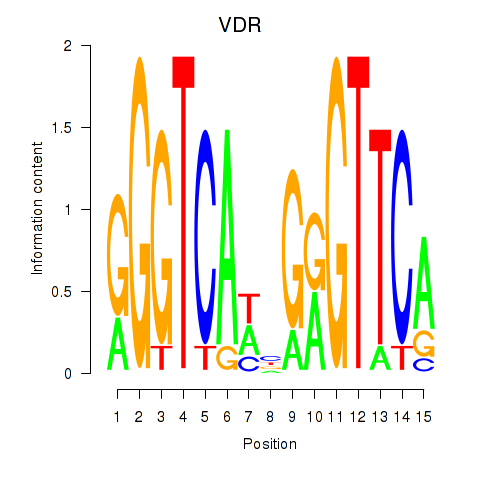
Transcription factors associated with VDR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VDR
|
ENSG00000111424.6 | vitamin D receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VDR | hg19_v2_chr12_-_48298785_48298828 | 0.41 | 2.3e-02 | Click! |
Activity profile of VDR motif
Sorted Z-values of VDR motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_102928009 | 4.28 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr16_-_2770216 | 2.69 |
ENST00000302641.3
|
PRSS27
|
protease, serine 27 |
| chr2_-_235405168 | 2.67 |
ENST00000339728.3
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr5_-_39270725 | 2.60 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr10_-_5541525 | 2.29 |
ENST00000380332.3
|
CALML5
|
calmodulin-like 5 |
| chr17_+_2699697 | 1.88 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr11_-_65667997 | 1.86 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr2_+_210517895 | 1.81 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr20_+_30327063 | 1.79 |
ENST00000300403.6
ENST00000340513.4 |
TPX2
|
TPX2, microtubule-associated |
| chr20_-_52790512 | 1.78 |
ENST00000216862.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr12_-_133050726 | 1.74 |
ENST00000595994.1
|
MUC8
|
mucin 8 |
| chr9_+_139839711 | 1.69 |
ENST00000224181.3
|
C8G
|
complement component 8, gamma polypeptide |
| chr11_-_28129656 | 1.66 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr11_-_65667884 | 1.59 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr1_+_17559776 | 1.58 |
ENST00000537499.1
ENST00000413717.2 ENST00000536552.1 |
PADI1
|
peptidyl arginine deiminase, type I |
| chr17_-_36347835 | 1.53 |
ENST00000519532.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr17_-_34807272 | 1.53 |
ENST00000535592.1
ENST00000394453.1 |
TBC1D3G
|
TBC1 domain family, member 3G |
| chr7_+_155090271 | 1.52 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr17_-_36348610 | 1.52 |
ENST00000339023.4
ENST00000354664.4 |
TBC1D3
|
TBC1 domain family, member 3 |
| chr17_-_34808047 | 1.52 |
ENST00000592614.1
ENST00000591542.1 ENST00000330458.7 ENST00000341264.6 ENST00000592987.1 ENST00000400684.4 |
TBC1D3G
TBC1D3H
|
TBC1 domain family, member 3G TBC1 domain family, member 3H |
| chr11_-_61582579 | 1.51 |
ENST00000539419.1
ENST00000545245.1 ENST00000545405.1 ENST00000542506.1 |
FADS1
|
fatty acid desaturase 1 |
| chr17_-_34756219 | 1.51 |
ENST00000451448.2
ENST00000394359.3 |
TBC1D3C
TBC1D3H
|
TBC1 domain family, member 3C TBC1 domain family, member 3H |
| chr7_+_143701022 | 1.43 |
ENST00000408922.2
|
OR6B1
|
olfactory receptor, family 6, subfamily B, member 1 |
| chr17_+_36284791 | 1.41 |
ENST00000505415.1
|
TBC1D3F
|
TBC1 domain family, member 3F |
| chr14_+_94640633 | 1.35 |
ENST00000304338.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr1_-_75198940 | 1.25 |
ENST00000417775.1
|
CRYZ
|
crystallin, zeta (quinone reductase) |
| chr16_+_2479390 | 1.23 |
ENST00000397066.4
|
CCNF
|
cyclin F |
| chr12_+_110011571 | 1.20 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr9_+_131644781 | 1.17 |
ENST00000259324.5
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr5_-_147211190 | 1.15 |
ENST00000510027.2
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr9_+_139839686 | 1.15 |
ENST00000371634.2
|
C8G
|
complement component 8, gamma polypeptide |
| chr8_+_26371763 | 1.09 |
ENST00000521913.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr1_-_221915418 | 1.09 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr19_+_38826415 | 1.08 |
ENST00000410018.1
ENST00000409235.3 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
| chr5_-_147211226 | 1.07 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr8_-_145028013 | 1.06 |
ENST00000354958.2
|
PLEC
|
plectin |
| chr4_+_142557771 | 1.04 |
ENST00000514653.1
|
IL15
|
interleukin 15 |
| chr10_+_54074033 | 1.00 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr8_+_95653427 | 1.00 |
ENST00000454170.2
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr10_+_80008505 | 0.99 |
ENST00000434974.1
ENST00000423770.1 ENST00000432742.1 |
LINC00856
|
long intergenic non-protein coding RNA 856 |
| chr11_-_64527425 | 0.98 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr17_-_34591208 | 0.95 |
ENST00000336331.5
|
TBC1D3C
|
TBC1 domain family, member 3C |
| chr4_-_103266626 | 0.93 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr8_-_22926526 | 0.92 |
ENST00000347739.3
ENST00000542226.1 |
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b |
| chr2_-_85625857 | 0.92 |
ENST00000453973.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chrX_-_65253506 | 0.92 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr19_-_16045665 | 0.92 |
ENST00000248041.8
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr17_-_43487741 | 0.87 |
ENST00000455881.1
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr19_-_16045619 | 0.87 |
ENST00000402119.4
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr12_+_57857475 | 0.85 |
ENST00000528467.1
|
GLI1
|
GLI family zinc finger 1 |
| chr1_+_153232160 | 0.85 |
ENST00000368742.3
|
LOR
|
loricrin |
| chr8_+_95653302 | 0.85 |
ENST00000423620.2
ENST00000433389.2 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr11_+_73882144 | 0.83 |
ENST00000328257.8
|
PPME1
|
protein phosphatase methylesterase 1 |
| chr8_+_95653373 | 0.82 |
ENST00000358397.5
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr13_+_114321463 | 0.82 |
ENST00000335678.6
|
GRK1
|
G protein-coupled receptor kinase 1 |
| chr1_-_38218577 | 0.80 |
ENST00000540011.1
|
EPHA10
|
EPH receptor A10 |
| chr11_+_73882311 | 0.76 |
ENST00000398427.4
ENST00000544401.1 |
PPME1
|
protein phosphatase methylesterase 1 |
| chr10_-_33623826 | 0.75 |
ENST00000374867.2
|
NRP1
|
neuropilin 1 |
| chr19_-_35992780 | 0.75 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr12_-_96390063 | 0.74 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr11_+_57365150 | 0.73 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr10_-_76995769 | 0.73 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr11_+_6624970 | 0.72 |
ENST00000420936.2
ENST00000528995.1 |
ILK
|
integrin-linked kinase |
| chr8_-_13134045 | 0.71 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr1_-_6550625 | 0.69 |
ENST00000377725.1
ENST00000340850.5 |
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr12_+_10658489 | 0.69 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr10_-_33623564 | 0.69 |
ENST00000374875.1
ENST00000374822.4 |
NRP1
|
neuropilin 1 |
| chr10_-_33624002 | 0.69 |
ENST00000432372.2
|
NRP1
|
neuropilin 1 |
| chr1_+_3816936 | 0.68 |
ENST00000413332.1
ENST00000442673.1 ENST00000439488.1 |
RP13-15E13.1
|
long intergenic non-protein coding RNA 1134 |
| chr7_+_65552756 | 0.67 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr21_-_47575481 | 0.67 |
ENST00000291670.5
ENST00000397748.1 ENST00000359679.2 ENST00000355384.2 ENST00000397746.3 ENST00000397743.1 |
FTCD
|
formimidoyltransferase cyclodeaminase |
| chr17_-_64225508 | 0.67 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr16_-_67217844 | 0.66 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr4_-_103266219 | 0.65 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr11_+_6625046 | 0.65 |
ENST00000396751.2
|
ILK
|
integrin-linked kinase |
| chr6_+_24775153 | 0.65 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr11_+_6624955 | 0.65 |
ENST00000299421.4
ENST00000537806.1 |
ILK
|
integrin-linked kinase |
| chr10_+_5454505 | 0.64 |
ENST00000355029.4
|
NET1
|
neuroepithelial cell transforming 1 |
| chr6_+_30525051 | 0.64 |
ENST00000376557.3
|
PRR3
|
proline rich 3 |
| chr11_+_28129795 | 0.64 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr19_+_1407517 | 0.63 |
ENST00000336761.6
ENST00000233078.4 |
DAZAP1
|
DAZ associated protein 1 |
| chr3_+_111578131 | 0.63 |
ENST00000498699.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr12_+_79439405 | 0.63 |
ENST00000552744.1
|
SYT1
|
synaptotagmin I |
| chr1_-_67519782 | 0.62 |
ENST00000235345.5
|
SLC35D1
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1 |
| chr8_-_22926623 | 0.62 |
ENST00000276431.4
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b |
| chr19_-_19932501 | 0.62 |
ENST00000540806.2
ENST00000590766.1 ENST00000587452.1 ENST00000545006.1 ENST00000590319.1 ENST00000587461.1 ENST00000450683.2 ENST00000443905.2 ENST00000590274.1 |
ZNF506
CTC-559E9.4
|
zinc finger protein 506 CTC-559E9.4 |
| chr17_-_43487780 | 0.61 |
ENST00000532038.1
ENST00000528677.1 |
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr17_-_46716647 | 0.60 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr6_+_30524663 | 0.60 |
ENST00000376560.3
|
PRR3
|
proline rich 3 |
| chr8_+_10383039 | 0.59 |
ENST00000328655.3
ENST00000522210.1 |
PRSS55
|
protease, serine, 55 |
| chr1_-_19426149 | 0.59 |
ENST00000429347.2
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr9_-_115095123 | 0.58 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr16_+_31119615 | 0.58 |
ENST00000394950.3
ENST00000287507.3 ENST00000219794.6 ENST00000561755.1 |
BCKDK
|
branched chain ketoacid dehydrogenase kinase |
| chr6_-_30524951 | 0.57 |
ENST00000376621.3
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr7_-_36764142 | 0.54 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chrX_+_153813407 | 0.54 |
ENST00000443287.2
ENST00000333128.3 |
CTAG1A
|
cancer/testis antigen 1A |
| chr17_+_78388959 | 0.52 |
ENST00000518137.1
ENST00000520367.1 ENST00000523999.1 ENST00000323854.5 ENST00000522751.1 |
ENDOV
|
endonuclease V |
| chr9_+_71789133 | 0.52 |
ENST00000348208.4
ENST00000265384.7 |
TJP2
|
tight junction protein 2 |
| chrX_+_43515467 | 0.52 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr7_-_22539771 | 0.51 |
ENST00000406890.2
ENST00000424363.1 |
STEAP1B
|
STEAP family member 1B |
| chr12_+_57610562 | 0.51 |
ENST00000349394.5
|
NXPH4
|
neurexophilin 4 |
| chr9_+_138391805 | 0.50 |
ENST00000371785.1
|
MRPS2
|
mitochondrial ribosomal protein S2 |
| chr2_+_198557830 | 0.50 |
ENST00000409845.1
|
AC011997.1
|
Uncharacterized protein |
| chr19_+_50270219 | 0.49 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr15_-_99057551 | 0.46 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr12_-_114404111 | 0.45 |
ENST00000545145.2
ENST00000392561.3 ENST00000261741.5 |
RBM19
|
RNA binding motif protein 19 |
| chrX_-_153881842 | 0.45 |
ENST00000369585.3
ENST00000247306.4 |
CTAG2
|
cancer/testis antigen 2 |
| chr2_+_220363579 | 0.45 |
ENST00000313597.5
ENST00000373917.3 ENST00000358215.3 ENST00000373908.1 ENST00000455657.1 ENST00000435316.1 ENST00000341142.3 |
GMPPA
|
GDP-mannose pyrophosphorylase A |
| chr4_+_158142750 | 0.45 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr9_+_130213774 | 0.45 |
ENST00000373324.4
ENST00000323301.4 |
LRSAM1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr14_+_22984601 | 0.45 |
ENST00000390509.1
|
TRAJ28
|
T cell receptor alpha joining 28 |
| chr2_+_87769459 | 0.45 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr3_+_54157480 | 0.44 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr3_-_122283100 | 0.43 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr5_-_82969405 | 0.43 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr16_-_2260834 | 0.43 |
ENST00000562360.1
ENST00000566018.1 |
BRICD5
|
BRICHOS domain containing 5 |
| chr13_-_33859819 | 0.43 |
ENST00000336934.5
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr2_+_108905095 | 0.43 |
ENST00000251481.6
ENST00000326853.5 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr11_-_104817919 | 0.43 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr20_+_55904815 | 0.41 |
ENST00000371263.3
ENST00000345868.4 ENST00000371260.4 ENST00000418127.1 |
SPO11
|
SPO11 meiotic protein covalently bound to DSB |
| chr14_-_75330537 | 0.41 |
ENST00000556084.2
ENST00000556489.2 ENST00000445876.1 |
PROX2
|
prospero homeobox 2 |
| chr14_+_105212297 | 0.41 |
ENST00000556623.1
ENST00000555674.1 |
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr14_-_24806588 | 0.40 |
ENST00000555591.1
ENST00000554569.1 |
RP11-934B9.3
RIPK3
|
Uncharacterized protein receptor-interacting serine-threonine kinase 3 |
| chr12_+_48876275 | 0.40 |
ENST00000314014.2
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr15_+_83776324 | 0.40 |
ENST00000379390.6
ENST00000379386.4 ENST00000565774.1 ENST00000565982.1 |
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr4_-_164534657 | 0.39 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr7_-_36764004 | 0.39 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr6_+_167704838 | 0.39 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr13_-_38172863 | 0.39 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr1_+_26348259 | 0.38 |
ENST00000374280.3
|
EXTL1
|
exostosin-like glycosyltransferase 1 |
| chr17_+_53828333 | 0.38 |
ENST00000268896.5
|
PCTP
|
phosphatidylcholine transfer protein |
| chr11_-_111794446 | 0.36 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr17_+_79213039 | 0.36 |
ENST00000431388.2
|
C17orf89
|
chromosome 17 open reading frame 89 |
| chr10_-_18940501 | 0.35 |
ENST00000377304.4
|
NSUN6
|
NOP2/Sun domain family, member 6 |
| chr15_+_41851211 | 0.35 |
ENST00000263798.3
|
TYRO3
|
TYRO3 protein tyrosine kinase |
| chr7_+_155089486 | 0.35 |
ENST00000340368.4
ENST00000344756.4 ENST00000425172.1 ENST00000342407.5 |
INSIG1
|
insulin induced gene 1 |
| chr20_+_30028322 | 0.35 |
ENST00000376309.3
|
DEFB123
|
defensin, beta 123 |
| chr19_-_40331345 | 0.35 |
ENST00000597224.1
|
FBL
|
fibrillarin |
| chr17_+_53828381 | 0.34 |
ENST00000576183.1
|
PCTP
|
phosphatidylcholine transfer protein |
| chr1_+_47137445 | 0.34 |
ENST00000569393.1
ENST00000334122.4 ENST00000415500.1 |
TEX38
|
testis expressed 38 |
| chr13_+_57721622 | 0.34 |
ENST00000377930.1
|
PRR20B
|
proline rich 20B |
| chr7_-_151433393 | 0.34 |
ENST00000492843.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr22_-_17680472 | 0.34 |
ENST00000330232.4
|
CECR1
|
cat eye syndrome chromosome region, candidate 1 |
| chr1_+_47137544 | 0.33 |
ENST00000564373.1
|
TEX38
|
testis expressed 38 |
| chr20_+_58630972 | 0.33 |
ENST00000313426.1
|
C20orf197
|
chromosome 20 open reading frame 197 |
| chr7_-_151433342 | 0.33 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr11_+_18433840 | 0.33 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr14_-_20801427 | 0.33 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr6_+_167704798 | 0.33 |
ENST00000230256.3
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr15_+_75940218 | 0.33 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr2_+_11752379 | 0.32 |
ENST00000396123.1
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr14_-_106781017 | 0.32 |
ENST00000390612.2
|
IGHV4-28
|
immunoglobulin heavy variable 4-28 |
| chr12_-_71512027 | 0.32 |
ENST00000549357.1
|
CTD-2021H9.3
|
Uncharacterized protein |
| chr1_-_205325850 | 0.31 |
ENST00000537168.1
|
KLHDC8A
|
kelch domain containing 8A |
| chr18_-_44544607 | 0.30 |
ENST00000591973.2
|
TCEB3CL2
|
transcription elongation factor B polypeptide 3C-like 2 |
| chr2_-_202483867 | 0.30 |
ENST00000439802.1
ENST00000286195.3 ENST00000439140.1 ENST00000450242.1 |
ALS2CR11
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11 |
| chr12_-_96390108 | 0.30 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr16_+_1128781 | 0.29 |
ENST00000293897.4
ENST00000562758.1 |
SSTR5
|
somatostatin receptor 5 |
| chr16_-_31106048 | 0.29 |
ENST00000300851.6
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr11_+_124609823 | 0.29 |
ENST00000412681.2
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chr16_-_31106211 | 0.29 |
ENST00000532364.1
ENST00000529564.1 ENST00000319788.7 ENST00000354895.4 ENST00000394975.2 |
RP11-196G11.1
VKORC1
|
Uncharacterized protein vitamin K epoxide reductase complex, subunit 1 |
| chr1_-_200992827 | 0.28 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr8_+_9046503 | 0.28 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr12_-_109221160 | 0.28 |
ENST00000326470.5
|
SSH1
|
slingshot protein phosphatase 1 |
| chr18_-_44556449 | 0.28 |
ENST00000330682.2
|
TCEB3C
|
transcription elongation factor B polypeptide 3C (elongin A3) |
| chr2_+_176957619 | 0.28 |
ENST00000392539.3
|
HOXD13
|
homeobox D13 |
| chr19_-_49122384 | 0.28 |
ENST00000550973.1
ENST00000550645.1 ENST00000549273.1 ENST00000552588.1 |
RPL18
|
ribosomal protein L18 |
| chr9_+_131038425 | 0.27 |
ENST00000320188.5
ENST00000608796.1 ENST00000419867.2 ENST00000418976.1 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr22_+_50624323 | 0.27 |
ENST00000380909.4
ENST00000303434.4 |
TRABD
|
TraB domain containing |
| chr12_-_6579833 | 0.27 |
ENST00000396308.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr1_+_175036966 | 0.26 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chr14_-_91884150 | 0.26 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr2_+_108905325 | 0.26 |
ENST00000438339.1
ENST00000409880.1 ENST00000437390.2 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr6_+_46661575 | 0.26 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr6_+_56819895 | 0.26 |
ENST00000370748.3
|
BEND6
|
BEN domain containing 6 |
| chr10_+_104154229 | 0.25 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr16_+_640201 | 0.24 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr9_+_125376948 | 0.24 |
ENST00000297913.2
|
OR1Q1
|
olfactory receptor, family 1, subfamily Q, member 1 |
| chr1_+_24069952 | 0.24 |
ENST00000609199.1
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) |
| chr16_-_31105870 | 0.23 |
ENST00000394971.3
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr12_+_27091426 | 0.23 |
ENST00000546072.1
ENST00000327214.5 |
FGFR1OP2
|
FGFR1 oncogene partner 2 |
| chr16_-_31228680 | 0.23 |
ENST00000302964.3
|
PYDC1
|
PYD (pyrin domain) containing 1 |
| chr14_-_54908043 | 0.23 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr11_-_104840093 | 0.23 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr11_+_65154070 | 0.22 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr13_+_47127322 | 0.22 |
ENST00000389798.3
|
LRCH1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr3_-_45838011 | 0.22 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr17_-_79805146 | 0.22 |
ENST00000415593.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr19_-_54804173 | 0.22 |
ENST00000391744.3
ENST00000251390.3 |
LILRA3
|
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 |
| chr8_+_12809093 | 0.21 |
ENST00000528753.2
|
KIAA1456
|
KIAA1456 |
| chr19_-_13261160 | 0.21 |
ENST00000343587.5
ENST00000591197.1 |
STX10
|
syntaxin 10 |
| chr14_-_102771462 | 0.21 |
ENST00000522874.1
|
MOK
|
MOK protein kinase |
| chr9_+_111624577 | 0.21 |
ENST00000333999.3
|
ACTL7A
|
actin-like 7A |
| chr19_-_13261090 | 0.21 |
ENST00000588848.1
|
STX10
|
syntaxin 10 |
| chr16_+_28834303 | 0.20 |
ENST00000340394.8
ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L
|
ataxin 2-like |
| chr12_+_3069037 | 0.20 |
ENST00000397122.2
|
TEAD4
|
TEA domain family member 4 |
| chr14_-_102771516 | 0.20 |
ENST00000524214.1
ENST00000193029.6 ENST00000361847.2 |
MOK
|
MOK protein kinase |
| chr1_+_154474689 | 0.20 |
ENST00000368482.4
|
TDRD10
|
tudor domain containing 10 |
| chr11_+_113779704 | 0.20 |
ENST00000537778.1
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr18_-_44550534 | 0.20 |
ENST00000451265.1
|
TCEB3CL
|
transcription elongation factor B polypeptide 3C-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of VDR
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.6 | 1.9 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.6 | 1.8 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.6 | 3.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.4 | 1.8 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.4 | 2.1 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.4 | 1.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.4 | 2.2 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.3 | 1.0 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.3 | 1.7 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.3 | 1.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.3 | 1.6 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 1.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.2 | 0.6 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 0.9 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.2 | 0.5 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 1.6 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.4 | GO:0042138 | meiotic DNA double-strand break processing(GO:0000706) meiotic DNA double-strand break formation(GO:0042138) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.4 | GO:0035698 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.1 | 1.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.8 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.4 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 1.7 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.8 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.3 | GO:0034963 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.1 | 0.7 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.6 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 0.3 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 10.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 2.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.4 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.4 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.1 | 0.4 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 1.5 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 0.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.4 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 1.5 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) linoleic acid metabolic process(GO:0043651) |
| 0.1 | 1.2 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.1 | 2.0 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 0.7 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 1.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.5 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.2 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 2.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.8 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.3 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.3 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.1 | 0.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 1.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.5 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 2.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.9 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0044828 | positive regulation of translational initiation in response to stress(GO:0032058) negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.3 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 1.8 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 1.6 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.6 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 1.0 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.8 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.3 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.7 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 1.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 1.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.1 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 1.8 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.5 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 2.0 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.0 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.0 | GO:0061744 | psychomotor behavior(GO:0036343) motor behavior(GO:0061744) |
| 0.0 | 0.3 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 | 1.9 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.5 | 1.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 2.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 2.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 1.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.6 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 1.6 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.4 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 0.5 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.7 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.7 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 3.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.3 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 2.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.8 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 3.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.8 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 4.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0070761 | PCAF complex(GO:0000125) pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 1.6 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 1.4 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 4.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.3 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 1.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 2.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.4 | 1.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.4 | 1.8 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.3 | 1.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 0.9 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.3 | 0.8 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.3 | 1.5 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.2 | 1.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.2 | 1.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 1.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 1.5 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.2 | 2.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 0.8 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.4 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.4 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 2.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.3 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 0.6 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 1.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.6 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 1.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 1.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.5 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.7 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 2.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 9.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.0 | 0.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 1.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.4 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 1.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 1.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.8 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0005165 | neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 1.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 2.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 3.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 1.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |