Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
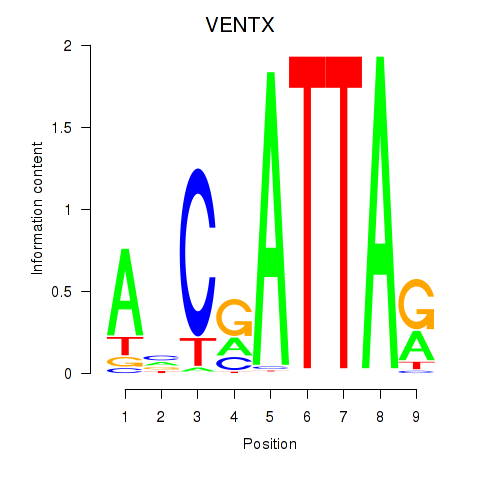
Results for VENTX
Z-value: 0.34
Transcription factors associated with VENTX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VENTX
|
ENSG00000151650.7 | VENT homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VENTX | hg19_v2_chr10_+_135050908_135050908 | -0.08 | 6.8e-01 | Click! |
Activity profile of VENTX motif
Sorted Z-values of VENTX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_67517716 | 0.72 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr12_+_107712173 | 0.64 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr12_-_89746173 | 0.46 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chrX_-_153602991 | 0.41 |
ENST00000369850.3
ENST00000422373.1 |
FLNA
|
filamin A, alpha |
| chr2_+_171034646 | 0.36 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr1_+_203651937 | 0.35 |
ENST00000341360.2
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr6_-_138820624 | 0.28 |
ENST00000343505.5
|
NHSL1
|
NHS-like 1 |
| chr3_-_57260377 | 0.27 |
ENST00000495160.2
|
HESX1
|
HESX homeobox 1 |
| chr2_-_163008903 | 0.26 |
ENST00000418842.2
ENST00000375497.3 |
GCG
|
glucagon |
| chr6_+_34204642 | 0.25 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr3_-_57233966 | 0.25 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr11_-_107729887 | 0.24 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr22_+_46476192 | 0.24 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr6_-_111927062 | 0.24 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr5_+_1225470 | 0.23 |
ENST00000324642.3
ENST00000296821.4 |
SLC6A18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr14_+_65171315 | 0.22 |
ENST00000394691.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr11_+_35198118 | 0.21 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr18_+_29171689 | 0.21 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr1_-_95391315 | 0.20 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr16_+_69984810 | 0.19 |
ENST00000393701.2
ENST00000568461.1 |
CLEC18A
|
C-type lectin domain family 18, member A |
| chr14_+_65453432 | 0.19 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr6_+_12290586 | 0.19 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr16_+_70207686 | 0.18 |
ENST00000541793.2
ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18, member C |
| chr16_+_69985083 | 0.18 |
ENST00000288040.6
ENST00000449317.2 |
CLEC18A
|
C-type lectin domain family 18, member A |
| chr1_+_226411319 | 0.18 |
ENST00000542034.1
ENST00000366810.5 |
MIXL1
|
Mix paired-like homeobox |
| chr17_-_39222131 | 0.18 |
ENST00000394015.2
|
KRTAP2-4
|
keratin associated protein 2-4 |
| chr21_+_39644395 | 0.17 |
ENST00000398934.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr12_-_57328187 | 0.17 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr6_+_37897735 | 0.17 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr6_-_66417107 | 0.16 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr10_-_56560939 | 0.16 |
ENST00000373955.1
|
PCDH15
|
protocadherin-related 15 |
| chrX_+_130192318 | 0.15 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr10_+_24738355 | 0.15 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr8_+_26150628 | 0.15 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr14_+_65171099 | 0.15 |
ENST00000247226.7
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr9_+_116225999 | 0.15 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr4_+_169013666 | 0.14 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr10_+_114043493 | 0.14 |
ENST00000369422.3
|
TECTB
|
tectorin beta |
| chr6_-_111927449 | 0.12 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr4_+_71587669 | 0.12 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr17_-_7307358 | 0.12 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr17_+_43239191 | 0.12 |
ENST00000589230.1
|
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr12_+_8666126 | 0.12 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr1_+_153746683 | 0.12 |
ENST00000271857.2
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr2_+_226265364 | 0.12 |
ENST00000272907.6
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr9_+_136501478 | 0.11 |
ENST00000393056.2
ENST00000263611.2 |
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr2_-_165698521 | 0.11 |
ENST00000409184.3
ENST00000392717.2 ENST00000456693.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr2_-_165698662 | 0.11 |
ENST00000194871.6
ENST00000445474.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr6_-_32157947 | 0.11 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr6_+_43968306 | 0.11 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr6_+_101847105 | 0.11 |
ENST00000369137.3
ENST00000318991.6 |
GRIK2
|
glutamate receptor, ionotropic, kainate 2 |
| chr18_+_59000815 | 0.11 |
ENST00000262717.4
|
CDH20
|
cadherin 20, type 2 |
| chr3_+_130569429 | 0.11 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr2_+_159825143 | 0.10 |
ENST00000454300.1
ENST00000263635.6 |
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chrX_+_130192216 | 0.10 |
ENST00000276211.5
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr4_+_119200215 | 0.10 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr11_-_3859089 | 0.10 |
ENST00000396979.1
|
RHOG
|
ras homolog family member G |
| chrX_+_106163626 | 0.10 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr12_+_111471828 | 0.09 |
ENST00000261726.6
|
CUX2
|
cut-like homeobox 2 |
| chr11_-_11374904 | 0.09 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr3_+_156799587 | 0.09 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr9_+_133539981 | 0.09 |
ENST00000253008.2
|
PRDM12
|
PR domain containing 12 |
| chr7_+_18535346 | 0.09 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr1_+_41174988 | 0.09 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr1_-_102312517 | 0.09 |
ENST00000338858.5
|
OLFM3
|
olfactomedin 3 |
| chr6_+_114178512 | 0.08 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr16_-_49890016 | 0.08 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr4_+_71457970 | 0.08 |
ENST00000322937.6
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr8_-_7220490 | 0.08 |
ENST00000400078.2
|
ZNF705G
|
zinc finger protein 705G |
| chrX_+_105937068 | 0.08 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr2_-_164592497 | 0.08 |
ENST00000333129.3
ENST00000409634.1 |
FIGN
|
fidgetin |
| chrX_+_135730297 | 0.08 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr11_+_64018955 | 0.07 |
ENST00000279230.6
ENST00000540288.1 ENST00000325234.5 |
PLCB3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr17_+_47448102 | 0.07 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr1_+_153747746 | 0.07 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr10_+_74451883 | 0.07 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr5_-_82969405 | 0.07 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr1_+_43291220 | 0.07 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr7_-_82792215 | 0.06 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr1_+_78470530 | 0.06 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr12_-_53012343 | 0.06 |
ENST00000305748.3
|
KRT73
|
keratin 73 |
| chr12_+_25205446 | 0.06 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr11_+_46722368 | 0.06 |
ENST00000311764.2
|
ZNF408
|
zinc finger protein 408 |
| chr1_+_150337100 | 0.06 |
ENST00000401000.4
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr3_-_27764190 | 0.06 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr5_-_142780280 | 0.05 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr14_+_22670455 | 0.05 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr8_+_31496809 | 0.05 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr4_-_26492076 | 0.05 |
ENST00000295589.3
|
CCKAR
|
cholecystokinin A receptor |
| chr8_+_38831683 | 0.05 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr20_+_42136308 | 0.05 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr12_+_25205568 | 0.05 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chrX_+_135730373 | 0.05 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr4_-_46126093 | 0.05 |
ENST00000295452.4
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr1_-_114696472 | 0.04 |
ENST00000393296.1
ENST00000369547.1 ENST00000610222.1 |
SYT6
|
synaptotagmin VI |
| chr8_+_107738343 | 0.04 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr13_+_76378407 | 0.04 |
ENST00000447038.1
|
LMO7
|
LIM domain 7 |
| chr2_+_169757750 | 0.04 |
ENST00000375363.3
ENST00000429379.2 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase, catalytic, 2 |
| chr22_+_39101728 | 0.04 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr1_-_178840157 | 0.04 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr12_+_70219052 | 0.04 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr6_+_153552455 | 0.04 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr8_+_92261516 | 0.04 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr22_-_40929812 | 0.04 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr1_+_186265399 | 0.04 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr12_-_102591604 | 0.04 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr9_-_23825956 | 0.04 |
ENST00000397312.2
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr2_-_88285309 | 0.04 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr12_-_53994805 | 0.04 |
ENST00000328463.7
|
ATF7
|
activating transcription factor 7 |
| chr20_-_25371412 | 0.03 |
ENST00000339157.5
ENST00000376542.3 |
ABHD12
|
abhydrolase domain containing 12 |
| chr12_+_25205666 | 0.03 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chrY_-_13524717 | 0.03 |
ENST00000331172.6
|
SLC9B1P1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 pseudogene 1 |
| chr7_-_104909435 | 0.03 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr18_-_53804580 | 0.03 |
ENST00000590484.1
ENST00000589293.1 ENST00000587904.1 ENST00000591974.1 |
RP11-456O19.4
|
RP11-456O19.4 |
| chr9_+_117904097 | 0.03 |
ENST00000374016.1
|
DEC1
|
deleted in esophageal cancer 1 |
| chr17_-_10372875 | 0.03 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr18_-_21977748 | 0.03 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr11_-_111649015 | 0.03 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr17_+_59489112 | 0.03 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr6_+_167704838 | 0.03 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr7_-_22234381 | 0.03 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr14_-_20801427 | 0.03 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr12_-_6233828 | 0.03 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr16_+_14280564 | 0.03 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr16_+_73420942 | 0.02 |
ENST00000554640.1
ENST00000562661.1 ENST00000561875.1 |
RP11-140I24.1
|
RP11-140I24.1 |
| chr7_+_148287657 | 0.02 |
ENST00000307003.2
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chrX_-_130423200 | 0.02 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr4_+_130017268 | 0.02 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr2_+_87135076 | 0.02 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr14_-_82000140 | 0.02 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr3_+_9834227 | 0.02 |
ENST00000287613.7
ENST00000397261.3 |
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa |
| chr11_+_75526212 | 0.02 |
ENST00000356136.3
|
UVRAG
|
UV radiation resistance associated |
| chr18_-_77276057 | 0.02 |
ENST00000597412.1
|
AC018445.1
|
Uncharacterized protein |
| chr17_-_46716647 | 0.02 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr3_+_9834179 | 0.02 |
ENST00000498623.2
|
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa |
| chr12_-_52828147 | 0.02 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr4_+_71458012 | 0.02 |
ENST00000449493.2
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr16_+_14280742 | 0.02 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr13_-_26795840 | 0.02 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr8_+_107738240 | 0.02 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr6_-_90529418 | 0.02 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr6_+_167704798 | 0.02 |
ENST00000230256.3
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr20_-_49307897 | 0.01 |
ENST00000535356.1
|
FAM65C
|
family with sequence similarity 65, member C |
| chr13_+_76378357 | 0.01 |
ENST00000489941.2
ENST00000525373.1 |
LMO7
|
LIM domain 7 |
| chr1_-_232651312 | 0.01 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr8_-_116681221 | 0.01 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr6_-_9977801 | 0.01 |
ENST00000316020.6
ENST00000491508.1 |
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr7_+_139025875 | 0.01 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr19_+_48949030 | 0.01 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr4_-_138453559 | 0.01 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr5_-_79950775 | 0.01 |
ENST00000439211.2
|
DHFR
|
dihydrofolate reductase |
| chr7_+_129984630 | 0.01 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr5_-_130500922 | 0.01 |
ENST00000513012.1
ENST00000508488.1 ENST00000506908.1 ENST00000304043.5 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr1_+_62417957 | 0.01 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr20_-_50722183 | 0.01 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr2_-_203776864 | 0.00 |
ENST00000261015.4
|
WDR12
|
WD repeat domain 12 |
| chr1_+_152943122 | 0.00 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr10_-_56561022 | 0.00 |
ENST00000373965.2
ENST00000414778.1 ENST00000395438.1 ENST00000409834.1 ENST00000395445.1 ENST00000395446.1 ENST00000395442.1 ENST00000395440.1 ENST00000395432.2 ENST00000361849.3 ENST00000395433.1 ENST00000320301.6 ENST00000395430.1 ENST00000437009.1 |
PCDH15
|
protocadherin-related 15 |
| chr10_-_49813090 | 0.00 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chrX_-_139587225 | 0.00 |
ENST00000370536.2
|
SOX3
|
SRY (sex determining region Y)-box 3 |
| chr12_-_86650077 | 0.00 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr11_+_22359562 | 0.00 |
ENST00000263160.3
|
SLC17A6
|
solute carrier family 17 (vesicular glutamate transporter), member 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VENTX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.4 | GO:0033241 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.1 | 0.7 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.4 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 0.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.2 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.3 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.5 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.1 | 0.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.0 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |