Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
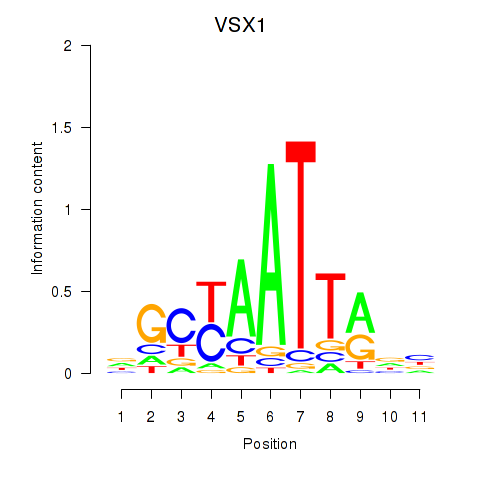
Results for VSX1
Z-value: 0.86
Transcription factors associated with VSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VSX1
|
ENSG00000100987.10 | visual system homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VSX1 | hg19_v2_chr20_-_25062767_25062779 | -0.08 | 6.7e-01 | Click! |
Activity profile of VSX1 motif
Sorted Z-values of VSX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_35938674 | 6.62 |
ENST00000397366.1
ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL
|
calcyphosine-like |
| chr12_+_7014064 | 4.59 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr12_+_7013897 | 4.58 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr3_-_197686847 | 4.20 |
ENST00000265239.6
|
IQCG
|
IQ motif containing G |
| chr17_-_1532106 | 3.48 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr1_-_36916066 | 3.05 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1 |
| chr12_+_7014126 | 2.63 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr1_-_36915880 | 2.52 |
ENST00000445843.3
|
OSCP1
|
organic solute carrier partner 1 |
| chr2_-_73460334 | 2.50 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr3_-_148939835 | 2.23 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr1_-_36916011 | 2.04 |
ENST00000356637.5
ENST00000354267.3 ENST00000235532.5 |
OSCP1
|
organic solute carrier partner 1 |
| chr19_+_32897009 | 1.84 |
ENST00000342179.5
ENST00000586427.1 |
DPY19L3
|
dpy-19-like 3 (C. elegans) |
| chr19_+_32896697 | 1.77 |
ENST00000586987.1
|
DPY19L3
|
dpy-19-like 3 (C. elegans) |
| chr5_-_54468974 | 1.76 |
ENST00000381375.2
ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B
|
cell division cycle 20B |
| chr11_-_26593779 | 1.71 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chrX_-_151999269 | 1.69 |
ENST00000370277.3
|
CETN2
|
centrin, EF-hand protein, 2 |
| chr4_-_138453559 | 1.66 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr11_-_33913708 | 1.63 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr11_-_95522907 | 1.61 |
ENST00000358780.5
ENST00000542135.1 |
FAM76B
|
family with sequence similarity 76, member B |
| chr3_-_47324008 | 1.48 |
ENST00000425853.1
|
KIF9
|
kinesin family member 9 |
| chr3_-_112693865 | 1.45 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr1_+_159750776 | 1.43 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr3_-_47324079 | 1.40 |
ENST00000352910.4
|
KIF9
|
kinesin family member 9 |
| chr1_-_23670817 | 1.40 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr19_+_50180317 | 1.34 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr11_-_95522639 | 1.31 |
ENST00000536839.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr3_-_112693759 | 1.30 |
ENST00000440122.2
ENST00000490004.1 |
CD200R1
|
CD200 receptor 1 |
| chr11_-_26593677 | 1.28 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr11_-_26593649 | 1.28 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr6_+_31783291 | 1.24 |
ENST00000375651.5
ENST00000608703.1 ENST00000458062.2 |
HSPA1A
|
heat shock 70kDa protein 1A |
| chr17_+_11501748 | 1.19 |
ENST00000262442.4
ENST00000579828.1 |
DNAH9
|
dynein, axonemal, heavy chain 9 |
| chr9_+_140135665 | 1.17 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr17_+_48172639 | 1.17 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chr3_-_47324242 | 1.11 |
ENST00000456548.1
ENST00000432493.1 ENST00000335044.2 ENST00000444589.2 |
KIF9
|
kinesin family member 9 |
| chr2_+_219135115 | 1.11 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr7_-_6048650 | 1.11 |
ENST00000382321.4
ENST00000406569.3 |
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr7_-_6048702 | 1.08 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr8_-_10512569 | 1.07 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr6_+_33422343 | 1.05 |
ENST00000395064.2
|
ZBTB9
|
zinc finger and BTB domain containing 9 |
| chr1_-_23670752 | 1.05 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr13_-_110438914 | 1.04 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr11_-_59383617 | 1.04 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr9_+_1050331 | 1.02 |
ENST00000382255.3
ENST00000382251.3 ENST00000412350.2 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr16_+_53133070 | 1.02 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr1_-_23670813 | 1.02 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr11_-_133826852 | 0.96 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr3_-_47324060 | 0.95 |
ENST00000452770.2
|
KIF9
|
kinesin family member 9 |
| chr8_-_29120580 | 0.94 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr20_+_54933971 | 0.93 |
ENST00000371384.3
ENST00000437418.1 |
FAM210B
|
family with sequence similarity 210, member B |
| chr2_-_217560248 | 0.92 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr1_-_169455169 | 0.92 |
ENST00000367804.4
ENST00000236137.5 |
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr19_+_11485333 | 0.90 |
ENST00000312423.2
|
SWSAP1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr2_+_10183651 | 0.90 |
ENST00000305883.1
|
KLF11
|
Kruppel-like factor 11 |
| chr9_+_131799213 | 0.90 |
ENST00000358369.4
ENST00000406926.2 ENST00000277475.5 ENST00000450073.1 |
FAM73B
|
family with sequence similarity 73, member B |
| chr17_-_42277203 | 0.89 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr5_+_148737562 | 0.88 |
ENST00000274569.4
|
PCYOX1L
|
prenylcysteine oxidase 1 like |
| chr4_-_141348789 | 0.86 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr11_-_122932730 | 0.85 |
ENST00000532182.1
ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr6_+_27833034 | 0.84 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr1_+_44115814 | 0.84 |
ENST00000372396.3
|
KDM4A
|
lysine (K)-specific demethylase 4A |
| chr6_+_36164487 | 0.82 |
ENST00000357641.6
|
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr20_+_60813535 | 0.81 |
ENST00000358053.2
ENST00000313733.3 ENST00000439951.2 |
OSBPL2
|
oxysterol binding protein-like 2 |
| chr11_-_3818688 | 0.81 |
ENST00000355260.3
ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98
|
nucleoporin 98kDa |
| chr11_+_57480046 | 0.80 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chr8_+_105235572 | 0.77 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr4_-_105416039 | 0.77 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr1_-_167905225 | 0.76 |
ENST00000367846.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr5_-_13944652 | 0.76 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr6_-_26271612 | 0.75 |
ENST00000305910.3
|
HIST1H3G
|
histone cluster 1, H3g |
| chr11_-_76155700 | 0.73 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr20_+_42086525 | 0.72 |
ENST00000244020.3
|
SRSF6
|
serine/arginine-rich splicing factor 6 |
| chr11_-_57479673 | 0.72 |
ENST00000337672.2
ENST00000431606.2 |
MED19
|
mediator complex subunit 19 |
| chr10_-_127505167 | 0.72 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr16_-_3350614 | 0.72 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr6_+_31795506 | 0.72 |
ENST00000375650.3
|
HSPA1B
|
heat shock 70kDa protein 1B |
| chr22_-_42486747 | 0.71 |
ENST00000602404.1
|
NDUFA6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa |
| chr18_-_71959159 | 0.70 |
ENST00000494131.2
ENST00000397914.4 ENST00000340533.4 |
CYB5A
|
cytochrome b5 type A (microsomal) |
| chr15_+_43803143 | 0.70 |
ENST00000382031.1
|
MAP1A
|
microtubule-associated protein 1A |
| chr3_-_195538728 | 0.70 |
ENST00000349607.4
ENST00000346145.4 |
MUC4
|
mucin 4, cell surface associated |
| chr3_-_186288097 | 0.70 |
ENST00000446782.1
|
TBCCD1
|
TBCC domain containing 1 |
| chr7_+_99933730 | 0.70 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr3_-_195538760 | 0.69 |
ENST00000475231.1
|
MUC4
|
mucin 4, cell surface associated |
| chr11_-_4629388 | 0.68 |
ENST00000526337.1
ENST00000300747.5 |
TRIM68
|
tripartite motif containing 68 |
| chr2_-_98280383 | 0.67 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr3_-_105587879 | 0.67 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr15_-_78526855 | 0.67 |
ENST00000541759.1
ENST00000558130.1 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr11_-_122933043 | 0.67 |
ENST00000534624.1
ENST00000453788.2 ENST00000527387.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr9_-_136933134 | 0.67 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr7_-_77427676 | 0.67 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr15_-_41522889 | 0.67 |
ENST00000458580.2
ENST00000314992.5 ENST00000558396.1 |
EXD1
|
exonuclease 3'-5' domain containing 1 |
| chr4_+_155702496 | 0.65 |
ENST00000510397.1
|
RBM46
|
RNA binding motif protein 46 |
| chr20_+_10415931 | 0.64 |
ENST00000334534.5
|
SLX4IP
|
SLX4 interacting protein |
| chr17_+_79981144 | 0.64 |
ENST00000306688.3
|
LRRC45
|
leucine rich repeat containing 45 |
| chr6_-_139613269 | 0.62 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr15_-_77363513 | 0.62 |
ENST00000267970.4
|
TSPAN3
|
tetraspanin 3 |
| chr11_-_76155618 | 0.62 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr15_-_37393406 | 0.61 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr11_-_124981475 | 0.61 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr14_-_78083112 | 0.60 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr1_+_152943122 | 0.59 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr10_-_50970322 | 0.58 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr3_+_152552685 | 0.57 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr19_+_50180507 | 0.57 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr18_-_61329118 | 0.56 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr1_-_45956800 | 0.56 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr18_-_45457478 | 0.56 |
ENST00000402690.2
ENST00000356825.4 |
SMAD2
|
SMAD family member 2 |
| chr19_+_19174795 | 0.55 |
ENST00000318596.7
|
SLC25A42
|
solute carrier family 25, member 42 |
| chr19_+_56652686 | 0.55 |
ENST00000592949.1
|
ZNF444
|
zinc finger protein 444 |
| chr3_+_186288454 | 0.54 |
ENST00000265028.3
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr19_+_46498704 | 0.54 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr7_+_6048856 | 0.52 |
ENST00000223029.3
ENST00000400479.2 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr19_+_50180409 | 0.52 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr2_-_96874553 | 0.52 |
ENST00000337288.5
ENST00000443962.1 |
STARD7
|
StAR-related lipid transfer (START) domain containing 7 |
| chr1_-_6052463 | 0.52 |
ENST00000378156.4
|
NPHP4
|
nephronophthisis 4 |
| chr7_+_55433131 | 0.51 |
ENST00000254770.2
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr19_-_47137942 | 0.51 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr19_+_39881951 | 0.50 |
ENST00000315588.5
ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29
|
mediator complex subunit 29 |
| chr1_-_91487770 | 0.50 |
ENST00000337393.5
|
ZNF644
|
zinc finger protein 644 |
| chr2_-_227050079 | 0.50 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr2_+_172864490 | 0.50 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr16_-_30773372 | 0.50 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr3_+_197687071 | 0.49 |
ENST00000482695.1
ENST00000330198.4 ENST00000419117.1 ENST00000420910.2 ENST00000332636.5 |
LMLN
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr20_-_17662705 | 0.49 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr10_-_50970382 | 0.49 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr2_+_86947296 | 0.49 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr11_+_3819049 | 0.48 |
ENST00000396986.2
ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2
|
post-GPI attachment to proteins 2 |
| chr13_+_50018402 | 0.48 |
ENST00000354234.4
|
SETDB2
|
SET domain, bifurcated 2 |
| chr19_-_39421377 | 0.48 |
ENST00000430193.3
ENST00000600042.1 ENST00000221431.6 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chrX_-_77150911 | 0.48 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr2_-_71454185 | 0.48 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr15_+_34638066 | 0.48 |
ENST00000333756.4
|
NUTM1
|
NUT midline carcinoma, family member 1 |
| chr4_-_168155169 | 0.48 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr2_-_88927092 | 0.47 |
ENST00000303236.3
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr17_+_57232690 | 0.47 |
ENST00000262293.4
|
PRR11
|
proline rich 11 |
| chr13_-_50018140 | 0.47 |
ENST00000410043.1
ENST00000347776.5 |
CAB39L
|
calcium binding protein 39-like |
| chr16_-_4784128 | 0.47 |
ENST00000592711.1
ENST00000590147.1 ENST00000304283.4 ENST00000592190.1 ENST00000589065.1 ENST00000585773.1 ENST00000450067.2 ENST00000592698.1 ENST00000586166.1 ENST00000586605.1 ENST00000592421.1 |
ANKS3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr1_+_24286287 | 0.47 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr17_-_47841485 | 0.46 |
ENST00000506156.1
ENST00000240364.2 |
FAM117A
|
family with sequence similarity 117, member A |
| chr19_+_13001840 | 0.45 |
ENST00000222214.5
ENST00000589039.1 ENST00000591470.1 ENST00000457854.1 ENST00000422947.2 ENST00000588905.1 ENST00000587072.1 |
GCDH
|
glutaryl-CoA dehydrogenase |
| chr22_-_32651326 | 0.45 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr19_+_1103936 | 0.45 |
ENST00000354171.8
ENST00000589115.1 |
GPX4
|
glutathione peroxidase 4 |
| chr1_-_150669500 | 0.45 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr2_-_240322643 | 0.45 |
ENST00000345617.3
|
HDAC4
|
histone deacetylase 4 |
| chr5_+_141303373 | 0.44 |
ENST00000432126.2
ENST00000194118.4 |
KIAA0141
|
KIAA0141 |
| chr13_+_19756173 | 0.44 |
ENST00000382988.2
|
RP11-408E5.4
|
RP11-408E5.4 |
| chr3_+_179280668 | 0.44 |
ENST00000429709.2
ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A
|
actin-like 6A |
| chr5_+_53751445 | 0.43 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr19_+_41770269 | 0.43 |
ENST00000378215.4
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr16_-_58663720 | 0.43 |
ENST00000564557.1
ENST00000569240.1 ENST00000441024.2 ENST00000569020.1 ENST00000317147.5 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr7_-_137686791 | 0.42 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr7_-_44887620 | 0.42 |
ENST00000349299.3
ENST00000521529.1 ENST00000308153.4 ENST00000350771.3 ENST00000222690.6 ENST00000381124.5 ENST00000437072.1 ENST00000446531.1 |
H2AFV
|
H2A histone family, member V |
| chr19_+_41770349 | 0.42 |
ENST00000602130.1
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr9_+_136399929 | 0.42 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS-like 2 |
| chr18_-_21017817 | 0.42 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr1_-_23520755 | 0.42 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr1_+_6845384 | 0.41 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr3_-_105588231 | 0.41 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chrM_+_4431 | 0.41 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr17_-_39203519 | 0.40 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr4_-_87028478 | 0.40 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr19_-_49622348 | 0.40 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr19_+_45394477 | 0.40 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr1_-_11322551 | 0.40 |
ENST00000361445.4
|
MTOR
|
mechanistic target of rapamycin (serine/threonine kinase) |
| chr6_-_85474219 | 0.39 |
ENST00000369663.5
|
TBX18
|
T-box 18 |
| chr20_-_17662878 | 0.39 |
ENST00000377813.1
ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
| chr7_+_74072288 | 0.39 |
ENST00000443166.1
|
GTF2I
|
general transcription factor IIi |
| chr7_+_138145145 | 0.39 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr5_+_159343688 | 0.38 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr17_-_1733114 | 0.38 |
ENST00000305513.7
|
SMYD4
|
SET and MYND domain containing 4 |
| chr19_+_41770236 | 0.38 |
ENST00000392006.3
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr2_-_131099897 | 0.38 |
ENST00000409127.1
ENST00000437688.2 ENST00000259229.2 |
CCDC115
|
coiled-coil domain containing 115 |
| chr15_-_77363441 | 0.38 |
ENST00000346495.2
ENST00000424443.3 ENST00000561277.1 |
TSPAN3
|
tetraspanin 3 |
| chr14_+_24583836 | 0.38 |
ENST00000559115.1
ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chrX_+_130192318 | 0.37 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr13_-_41240717 | 0.37 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr11_+_67798090 | 0.37 |
ENST00000313468.5
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr12_-_49076002 | 0.37 |
ENST00000357861.3
ENST00000550347.1 ENST00000420613.2 ENST00000550931.1 ENST00000550870.1 |
KANSL2
|
KAT8 regulatory NSL complex subunit 2 |
| chr19_-_47616992 | 0.37 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr12_+_118573663 | 0.36 |
ENST00000261313.2
|
PEBP1
|
phosphatidylethanolamine binding protein 1 |
| chr5_+_98109322 | 0.36 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr19_-_6737576 | 0.36 |
ENST00000601716.1
ENST00000264080.7 |
GPR108
|
G protein-coupled receptor 108 |
| chrX_+_107334895 | 0.36 |
ENST00000372232.3
ENST00000345734.3 ENST00000372254.3 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr16_+_68119324 | 0.36 |
ENST00000349223.5
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr2_-_61697862 | 0.36 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr5_+_177631523 | 0.35 |
ENST00000506339.1
ENST00000355836.5 ENST00000514633.1 ENST00000515193.1 ENST00000506259.1 ENST00000504898.1 |
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr6_+_36165133 | 0.35 |
ENST00000446974.1
ENST00000454960.1 |
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr1_-_149900122 | 0.35 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr13_-_41593425 | 0.35 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr19_-_54663473 | 0.35 |
ENST00000222224.3
|
LENG1
|
leukocyte receptor cluster (LRC) member 1 |
| chr15_+_89631381 | 0.34 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr3_-_42003479 | 0.34 |
ENST00000420927.1
|
ULK4
|
unc-51 like kinase 4 |
| chr7_-_86849883 | 0.34 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr5_-_147286065 | 0.34 |
ENST00000318315.4
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr19_+_38893751 | 0.33 |
ENST00000588262.1
ENST00000252530.5 ENST00000343358.7 |
FAM98C
|
family with sequence similarity 98, member C |
| chr8_-_52811564 | 0.33 |
ENST00000522514.1
|
PCMTD1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr3_-_44519131 | 0.33 |
ENST00000425708.2
ENST00000396077.2 |
ZNF445
|
zinc finger protein 445 |
| chr1_-_45956822 | 0.33 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr3_-_61237050 | 0.33 |
ENST00000476844.1
ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT
|
fragile histidine triad |
| chr3_-_120461378 | 0.33 |
ENST00000273375.3
|
RABL3
|
RAB, member of RAS oncogene family-like 3 |
| chr5_+_177631497 | 0.33 |
ENST00000358344.3
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
Network of associatons between targets according to the STRING database.
First level regulatory network of VSX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.5 | 3.6 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 1.3 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.4 | 4.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.4 | 2.4 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.4 | 1.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.3 | 1.5 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.3 | 0.9 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.2 | 1.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 | 0.7 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.2 | 0.9 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.2 | 1.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 0.6 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.2 | 0.8 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.2 | 0.6 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.2 | 0.6 | GO:0080121 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.2 | 0.6 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.2 | 0.5 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.2 | 1.0 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.2 | 0.5 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 0.5 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.2 | 1.0 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 0.5 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.7 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.1 | 0.6 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.4 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 1.2 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.1 | 0.4 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.1 | 4.6 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.1 | 2.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.8 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 3.4 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.3 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.8 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.4 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.3 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.1 | 0.9 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.7 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.4 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.1 | 2.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 1.0 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 1.6 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 8.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 3.5 | GO:1902475 | neutral amino acid transport(GO:0015804) L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 1.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 1.3 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 0.2 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.7 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.2 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.1 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.3 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.4 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.1 | 0.5 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 1.7 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.1 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 3.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.0 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.6 | GO:0006568 | tryptophan metabolic process(GO:0006568) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.4 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.9 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.7 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 1.2 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:1902177 | positive regulation of sister chromatid cohesion(GO:0045876) positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.5 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 2.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 1.1 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.3 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0034182 | regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) |
| 0.0 | 0.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 1.0 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 1.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0072302 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.7 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.3 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.0 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.9 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.2 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 1.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 2.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.2 | 0.9 | GO:0097196 | Shu complex(GO:0097196) |
| 0.2 | 0.7 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 0.7 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 1.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.2 | 1.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 7.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.7 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 1.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.9 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.8 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.7 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.1 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 5.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 3.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 5.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 2.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 2.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 3.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.6 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 2.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.3 | 2.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.3 | 1.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.3 | 3.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 2.2 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.3 | 0.9 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.3 | 1.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 2.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 1.4 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.2 | 1.8 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 0.6 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 0.5 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 0.8 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 0.5 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.2 | 1.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 0.8 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.6 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 3.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.4 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 0.9 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 0.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 3.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 4.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.2 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 5.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.3 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.0 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 2.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.5 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 3.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 4.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 5.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 1.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 6.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 2.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |