Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
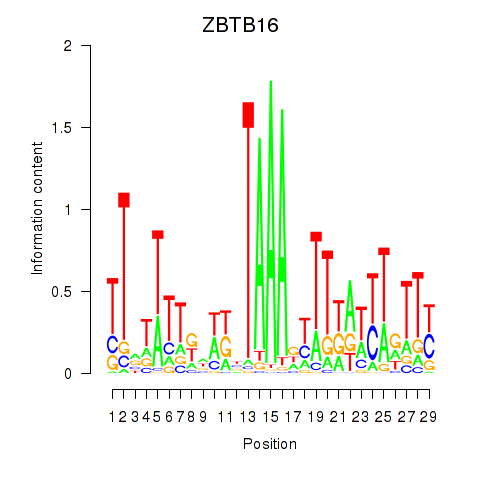
Results for ZBTB16
Z-value: 0.54
Transcription factors associated with ZBTB16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB16
|
ENSG00000109906.9 | zinc finger and BTB domain containing 16 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB16 | hg19_v2_chr11_+_113930291_113930339 | 0.18 | 3.5e-01 | Click! |
Activity profile of ZBTB16 motif
Sorted Z-values of ZBTB16 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_48082192 | 1.42 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr12_+_4385230 | 1.06 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr1_+_104159999 | 0.83 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr5_+_96212185 | 0.74 |
ENST00000379904.4
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr12_+_21207503 | 0.71 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr15_+_69365265 | 0.69 |
ENST00000415504.1
|
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr12_-_91539918 | 0.59 |
ENST00000548218.1
|
DCN
|
decorin |
| chr22_+_38203898 | 0.56 |
ENST00000323205.6
ENST00000248924.6 ENST00000445195.1 |
GCAT
|
glycine C-acetyltransferase |
| chr12_+_20968608 | 0.53 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr19_+_20959098 | 0.53 |
ENST00000360204.5
ENST00000594534.1 |
ZNF66
|
zinc finger protein 66 |
| chr4_+_74301880 | 0.52 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr17_+_61086917 | 0.49 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr3_-_120365866 | 0.47 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr5_-_55412774 | 0.46 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr10_-_61495760 | 0.46 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr8_+_11961898 | 0.45 |
ENST00000400085.3
|
ZNF705D
|
zinc finger protein 705D |
| chr20_-_18477862 | 0.43 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr19_+_10123925 | 0.36 |
ENST00000591589.1
ENST00000171214.1 |
RDH8
|
retinol dehydrogenase 8 (all-trans) |
| chr5_+_140220769 | 0.35 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr15_-_54051831 | 0.33 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr17_-_39341594 | 0.32 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr9_+_105757590 | 0.31 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr3_-_137893721 | 0.31 |
ENST00000505015.2
ENST00000260803.4 |
DBR1
|
debranching RNA lariats 1 |
| chr10_-_47239738 | 0.30 |
ENST00000413193.2
|
AGAP10
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr8_+_104831554 | 0.30 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr10_+_48189612 | 0.29 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr14_+_22458631 | 0.29 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr12_+_113682066 | 0.28 |
ENST00000392569.4
ENST00000552542.1 |
TPCN1
|
two pore segment channel 1 |
| chr16_-_18908196 | 0.28 |
ENST00000565324.1
ENST00000561947.1 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr1_+_12916941 | 0.28 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr18_-_52626622 | 0.28 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr13_+_77564795 | 0.26 |
ENST00000377453.3
|
CLN5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr12_-_113772835 | 0.26 |
ENST00000552014.1
ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr2_+_102618428 | 0.25 |
ENST00000457817.1
|
IL1R2
|
interleukin 1 receptor, type II |
| chr12_+_48166978 | 0.25 |
ENST00000442218.2
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr5_+_67586465 | 0.25 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr1_+_110993795 | 0.25 |
ENST00000271331.3
|
PROK1
|
prokineticin 1 |
| chr4_+_74606223 | 0.25 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr6_-_34639733 | 0.25 |
ENST00000374021.1
|
C6orf106
|
chromosome 6 open reading frame 106 |
| chr19_-_13617247 | 0.24 |
ENST00000573710.2
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr12_-_68696652 | 0.24 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr6_+_37897735 | 0.24 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr8_-_122653630 | 0.23 |
ENST00000303924.4
|
HAS2
|
hyaluronan synthase 2 |
| chr3_-_155524049 | 0.23 |
ENST00000534941.1
ENST00000340171.2 |
C3orf33
|
chromosome 3 open reading frame 33 |
| chr9_-_110540419 | 0.23 |
ENST00000398726.3
|
AL162389.1
|
Uncharacterized protein |
| chr19_+_21265028 | 0.23 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr16_-_18887627 | 0.22 |
ENST00000563235.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr19_-_56343353 | 0.22 |
ENST00000592953.1
ENST00000589093.1 |
NLRP11
|
NLR family, pyrin domain containing 11 |
| chr6_+_134758827 | 0.22 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr16_+_89984287 | 0.22 |
ENST00000555147.1
|
MC1R
|
melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) |
| chr2_-_89310012 | 0.21 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chrX_-_14047996 | 0.21 |
ENST00000380523.4
ENST00000398355.3 |
GEMIN8
|
gem (nuclear organelle) associated protein 8 |
| chr10_-_28623368 | 0.21 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_+_12851545 | 0.21 |
ENST00000332296.7
|
PRAMEF1
|
PRAME family member 1 |
| chr1_-_44818599 | 0.21 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr12_-_10007448 | 0.21 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr19_-_23869999 | 0.20 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr3_-_93692781 | 0.20 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr1_+_36335351 | 0.20 |
ENST00000373206.1
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr17_+_19314505 | 0.20 |
ENST00000461366.1
|
RNF112
|
ring finger protein 112 |
| chr7_-_107770794 | 0.20 |
ENST00000205386.4
ENST00000418464.1 ENST00000388781.3 ENST00000388780.3 ENST00000414450.2 |
LAMB4
|
laminin, beta 4 |
| chr16_-_70239683 | 0.20 |
ENST00000601706.1
|
AC009060.1
|
Uncharacterized protein |
| chr2_-_242556900 | 0.19 |
ENST00000402545.1
ENST00000402136.1 |
THAP4
|
THAP domain containing 4 |
| chr2_+_105050794 | 0.19 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr7_-_32534850 | 0.19 |
ENST00000409952.3
ENST00000409909.3 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr8_+_104831472 | 0.19 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr4_+_154622652 | 0.18 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr19_-_19932501 | 0.18 |
ENST00000540806.2
ENST00000590766.1 ENST00000587452.1 ENST00000545006.1 ENST00000590319.1 ENST00000587461.1 ENST00000450683.2 ENST00000443905.2 ENST00000590274.1 |
ZNF506
CTC-559E9.4
|
zinc finger protein 506 CTC-559E9.4 |
| chr7_-_38289173 | 0.18 |
ENST00000436911.2
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr3_+_40518599 | 0.18 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr20_-_33264886 | 0.17 |
ENST00000217446.3
ENST00000452740.2 ENST00000374820.2 |
PIGU
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr11_-_111649015 | 0.17 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr19_+_3762645 | 0.17 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr8_-_8318847 | 0.16 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr8_+_7801144 | 0.16 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
| chr9_-_21239978 | 0.16 |
ENST00000380222.2
|
IFNA14
|
interferon, alpha 14 |
| chr9_-_36400213 | 0.16 |
ENST00000259605.6
ENST00000353739.4 |
RNF38
|
ring finger protein 38 |
| chr5_+_173763250 | 0.16 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr6_+_27925019 | 0.15 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr6_-_138893661 | 0.15 |
ENST00000427025.2
|
NHSL1
|
NHS-like 1 |
| chr19_-_23869970 | 0.15 |
ENST00000601010.1
|
ZNF675
|
zinc finger protein 675 |
| chr21_-_35284635 | 0.15 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chr20_-_54580523 | 0.15 |
ENST00000064571.2
|
CBLN4
|
cerebellin 4 precursor |
| chrY_+_26997726 | 0.14 |
ENST00000382296.2
|
DAZ4
|
deleted in azoospermia 4 |
| chr15_-_34880646 | 0.14 |
ENST00000543376.1
|
GOLGA8A
|
golgin A8 family, member A |
| chr19_+_3762703 | 0.14 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr2_+_242289502 | 0.14 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr19_+_13135386 | 0.14 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr16_+_22501658 | 0.14 |
ENST00000415833.2
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr1_+_32608566 | 0.14 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr22_+_30821732 | 0.13 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr10_+_118083919 | 0.13 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr19_+_9296279 | 0.13 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr2_+_191334212 | 0.13 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr12_+_118454500 | 0.13 |
ENST00000537315.1
ENST00000229043.3 ENST00000484086.2 ENST00000420967.1 ENST00000454402.2 ENST00000392542.2 ENST00000535092.1 |
RFC5
|
replication factor C (activator 1) 5, 36.5kDa |
| chr14_+_52164820 | 0.13 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr7_+_112120908 | 0.13 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr8_-_74205851 | 0.13 |
ENST00000396467.1
|
RPL7
|
ribosomal protein L7 |
| chr18_+_39535152 | 0.12 |
ENST00000262039.4
ENST00000398870.3 ENST00000586545.1 |
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr19_-_53324884 | 0.12 |
ENST00000457749.2
ENST00000414252.2 ENST00000391783.2 |
ZNF28
|
zinc finger protein 28 |
| chr5_-_43397184 | 0.12 |
ENST00000513525.1
|
CCL28
|
chemokine (C-C motif) ligand 28 |
| chrX_-_24045303 | 0.12 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr18_+_7946941 | 0.12 |
ENST00000444013.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr5_-_24645078 | 0.12 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr4_-_90758118 | 0.12 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chrX_-_73834449 | 0.11 |
ENST00000332687.6
ENST00000349225.2 |
RLIM
|
ring finger protein, LIM domain interacting |
| chr3_+_57875738 | 0.11 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr16_-_4588822 | 0.11 |
ENST00000564828.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chr5_+_68860949 | 0.11 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr14_+_23067146 | 0.11 |
ENST00000428304.2
|
ABHD4
|
abhydrolase domain containing 4 |
| chr1_+_156095951 | 0.11 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr1_-_115292591 | 0.11 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr16_+_76587314 | 0.10 |
ENST00000563764.1
|
RP11-58C22.1
|
Uncharacterized protein |
| chr20_+_31407692 | 0.10 |
ENST00000375571.5
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1 |
| chrX_-_30885319 | 0.10 |
ENST00000378933.1
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr9_+_131062367 | 0.10 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr8_+_132952112 | 0.10 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr15_-_50406680 | 0.10 |
ENST00000559829.1
|
ATP8B4
|
ATPase, class I, type 8B, member 4 |
| chr22_+_30821784 | 0.10 |
ENST00000407550.3
|
MTFP1
|
mitochondrial fission process 1 |
| chr18_-_21017817 | 0.10 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chrX_+_71401526 | 0.10 |
ENST00000218432.5
ENST00000423432.2 ENST00000373669.2 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr15_-_54025300 | 0.10 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chrX_+_140982452 | 0.09 |
ENST00000544766.1
|
MAGEC3
|
melanoma antigen family C, 3 |
| chr9_-_116837249 | 0.09 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr12_-_75905374 | 0.09 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr15_-_55562479 | 0.09 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr10_+_62538089 | 0.09 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr8_-_130799134 | 0.09 |
ENST00000276708.4
|
GSDMC
|
gasdermin C |
| chr14_+_77228532 | 0.09 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr12_-_25150373 | 0.09 |
ENST00000549828.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr18_-_48351743 | 0.08 |
ENST00000588444.1
ENST00000256425.2 ENST00000428869.2 |
MRO
|
maestro |
| chr17_+_21729593 | 0.08 |
ENST00000581769.1
ENST00000584755.1 |
UBBP4
|
ubiquitin B pseudogene 4 |
| chr9_+_79074068 | 0.08 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr5_-_34919094 | 0.08 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr7_-_36764004 | 0.08 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr7_-_36764142 | 0.08 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr4_+_96012614 | 0.08 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr1_+_170633047 | 0.08 |
ENST00000239461.6
ENST00000497230.2 |
PRRX1
|
paired related homeobox 1 |
| chr4_+_106631966 | 0.08 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr7_-_104909435 | 0.08 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr18_-_53069419 | 0.08 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr14_+_39703112 | 0.08 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr12_+_32832134 | 0.08 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr10_+_47894023 | 0.08 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr15_-_55562582 | 0.07 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr22_+_29168652 | 0.07 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr14_+_22670455 | 0.07 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr19_+_21264980 | 0.07 |
ENST00000596053.1
ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714
|
zinc finger protein 714 |
| chr5_-_175395283 | 0.07 |
ENST00000513482.1
ENST00000265097.4 |
THOC3
|
THO complex 3 |
| chr18_+_3466248 | 0.07 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr17_-_37607497 | 0.07 |
ENST00000394287.3
ENST00000300651.6 |
MED1
|
mediator complex subunit 1 |
| chr20_-_31592113 | 0.07 |
ENST00000420875.1
ENST00000375519.2 ENST00000375523.3 ENST00000356173.3 |
SUN5
|
Sad1 and UNC84 domain containing 5 |
| chr11_+_67351213 | 0.07 |
ENST00000398603.1
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr3_+_184529948 | 0.07 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr5_+_180682720 | 0.07 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr12_+_32832203 | 0.07 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr10_+_5005445 | 0.07 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr19_+_36142147 | 0.07 |
ENST00000590618.1
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr10_+_47894572 | 0.07 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr6_+_42749759 | 0.07 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chr4_+_70796784 | 0.07 |
ENST00000246891.4
ENST00000444405.3 |
CSN1S1
|
casein alpha s1 |
| chr17_+_46908350 | 0.07 |
ENST00000258947.3
ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chr14_+_39703084 | 0.06 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr4_-_140223614 | 0.06 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr4_-_70518941 | 0.06 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr3_+_184529929 | 0.06 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr9_-_106087316 | 0.06 |
ENST00000411575.1
|
RP11-341A22.2
|
RP11-341A22.2 |
| chr2_-_73520667 | 0.06 |
ENST00000545030.1
ENST00000436467.2 |
EGR4
|
early growth response 4 |
| chr12_+_15699286 | 0.06 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr6_+_88117683 | 0.06 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr12_-_71533055 | 0.06 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr7_+_1727755 | 0.06 |
ENST00000424383.2
|
ELFN1
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chrX_+_118602363 | 0.06 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr22_+_24198890 | 0.05 |
ENST00000345044.6
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr1_+_76251879 | 0.05 |
ENST00000535300.1
ENST00000319942.3 |
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr2_+_131862872 | 0.05 |
ENST00000439822.2
|
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr3_+_51663407 | 0.05 |
ENST00000432863.1
ENST00000296477.3 |
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr10_+_104614008 | 0.05 |
ENST00000369883.3
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr4_-_90758227 | 0.05 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr10_+_104613980 | 0.05 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr16_+_24549014 | 0.05 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr12_+_133657461 | 0.05 |
ENST00000412146.2
ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140
|
zinc finger protein 140 |
| chr3_-_169487617 | 0.05 |
ENST00000330368.2
|
ACTRT3
|
actin-related protein T3 |
| chr4_-_140223670 | 0.05 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr18_+_11857439 | 0.05 |
ENST00000602628.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr22_+_24407642 | 0.05 |
ENST00000454754.1
ENST00000263119.5 |
CABIN1
|
calcineurin binding protein 1 |
| chr18_+_32398326 | 0.05 |
ENST00000269192.7
ENST00000591182.1 ENST00000597674.1 ENST00000556414.3 |
DTNA
|
dystrobrevin, alpha |
| chr19_-_29704448 | 0.05 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr3_-_133969673 | 0.05 |
ENST00000427044.2
|
RYK
|
receptor-like tyrosine kinase |
| chr4_-_99850243 | 0.05 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr8_+_67039278 | 0.05 |
ENST00000276573.7
ENST00000350034.4 |
TRIM55
|
tripartite motif containing 55 |
| chr10_-_75226166 | 0.05 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr8_+_7682694 | 0.05 |
ENST00000335186.2
|
DEFB106A
|
defensin, beta 106A |
| chr11_+_67351019 | 0.04 |
ENST00000398606.3
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr19_+_34895289 | 0.04 |
ENST00000246535.3
|
PDCD2L
|
programmed cell death 2-like |
| chr10_+_43916052 | 0.04 |
ENST00000442526.2
|
RP11-517P14.2
|
RP11-517P14.2 |
| chr19_-_45004556 | 0.04 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr12_+_53836339 | 0.04 |
ENST00000549135.1
|
PRR13
|
proline rich 13 |
| chr8_+_67687413 | 0.04 |
ENST00000521960.1
ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr11_+_5009424 | 0.04 |
ENST00000300762.1
|
MMP26
|
matrix metallopeptidase 26 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB16
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.2 | 0.6 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 0.2 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.4 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.5 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.7 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.9 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.2 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.2 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.0 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 1.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.2 | GO:0090037 | positive regulation of protein kinase A signaling(GO:0010739) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.3 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.3 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.0 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.6 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 1.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) mediator complex binding(GO:0036033) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |