Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for ZBTB3
Z-value: 0.53
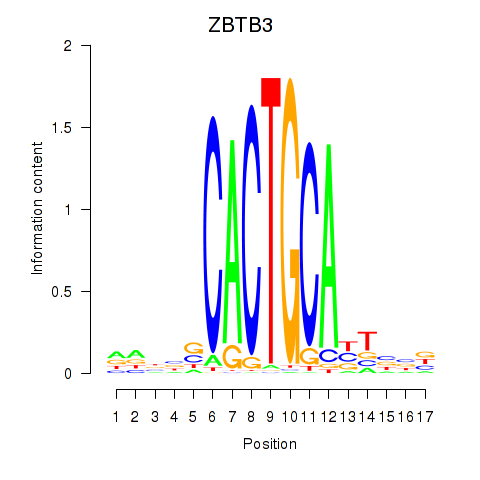
Transcription factors associated with ZBTB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB3
|
ENSG00000185670.7 | zinc finger and BTB domain containing 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB3 | hg19_v2_chr11_-_62521614_62521660 | 0.53 | 2.4e-03 | Click! |
Activity profile of ZBTB3 motif
Sorted Z-values of ZBTB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_152240819 | 2.05 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chrX_+_152338301 | 2.04 |
ENST00000453825.2
|
PNMA6A
|
paraneoplastic Ma antigen family member 6A |
| chr11_+_73661364 | 1.26 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr14_-_92413727 | 1.24 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr5_+_76506706 | 0.99 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr6_+_29274403 | 0.96 |
ENST00000377160.2
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1 |
| chr6_-_46138676 | 0.96 |
ENST00000371383.2
ENST00000230565.3 |
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) |
| chr2_-_154335300 | 0.95 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr14_-_92413353 | 0.91 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr14_-_92414055 | 0.91 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr16_-_68406161 | 0.86 |
ENST00000568373.1
ENST00000563226.1 |
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr1_+_211433275 | 0.71 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr19_+_49496705 | 0.68 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr16_-_19896220 | 0.63 |
ENST00000562469.1
ENST00000300571.2 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr19_+_49496782 | 0.60 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr1_+_110036699 | 0.60 |
ENST00000496961.1
ENST00000533024.1 ENST00000310611.4 ENST00000527072.1 ENST00000420578.2 ENST00000528785.1 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr7_+_23719749 | 0.58 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr5_+_140514782 | 0.58 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr1_+_110036728 | 0.57 |
ENST00000369868.3
ENST00000430195.2 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr17_-_17109579 | 0.52 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chr4_-_123542224 | 0.50 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr10_-_118032979 | 0.49 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr19_-_40023450 | 0.47 |
ENST00000326282.4
|
EID2B
|
EP300 interacting inhibitor of differentiation 2B |
| chr14_-_90085458 | 0.46 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr6_-_31939734 | 0.44 |
ENST00000375356.3
|
DXO
|
decapping exoribonuclease |
| chr5_-_82969405 | 0.43 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr4_-_7436671 | 0.41 |
ENST00000319098.4
|
PSAPL1
|
prosaposin-like 1 (gene/pseudogene) |
| chr1_+_160121356 | 0.37 |
ENST00000368081.4
|
ATP1A4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr17_+_33895090 | 0.36 |
ENST00000592381.1
|
RP11-1094M14.11
|
RP11-1094M14.11 |
| chr2_+_42396472 | 0.35 |
ENST00000318522.5
ENST00000402711.2 |
EML4
|
echinoderm microtubule associated protein like 4 |
| chr6_-_41122063 | 0.34 |
ENST00000426005.2
ENST00000437044.2 ENST00000373127.4 |
TREML1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr5_+_170288856 | 0.34 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr10_-_73533255 | 0.34 |
ENST00000394957.3
|
C10orf54
|
chromosome 10 open reading frame 54 |
| chr1_-_32801825 | 0.33 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr2_-_40679186 | 0.33 |
ENST00000406785.2
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr19_+_52848659 | 0.32 |
ENST00000327920.8
|
ZNF610
|
zinc finger protein 610 |
| chr6_+_30850697 | 0.32 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_+_42396574 | 0.32 |
ENST00000401738.3
|
EML4
|
echinoderm microtubule associated protein like 4 |
| chr11_-_67980744 | 0.31 |
ENST00000401547.2
ENST00000453170.1 ENST00000304363.4 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr2_+_241631262 | 0.30 |
ENST00000337801.4
ENST00000429564.1 |
AQP12A
|
aquaporin 12A |
| chr16_+_24266874 | 0.30 |
ENST00000005284.3
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr16_-_2185899 | 0.29 |
ENST00000262304.4
ENST00000423118.1 |
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr16_+_53164833 | 0.26 |
ENST00000564845.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_+_72002837 | 0.25 |
ENST00000377216.3
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr16_+_87636474 | 0.25 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr5_+_74807581 | 0.22 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chr12_-_114843889 | 0.22 |
ENST00000405440.2
|
TBX5
|
T-box 5 |
| chr6_+_42141029 | 0.22 |
ENST00000372958.1
|
GUCA1A
|
guanylate cyclase activator 1A (retina) |
| chr5_-_74807418 | 0.22 |
ENST00000405807.4
ENST00000261415.7 |
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr1_+_210001309 | 0.20 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr6_-_107436473 | 0.19 |
ENST00000369042.1
|
BEND3
|
BEN domain containing 3 |
| chr17_+_46018872 | 0.19 |
ENST00000583599.1
ENST00000434554.2 ENST00000225573.4 ENST00000544840.1 ENST00000534893.1 |
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr1_-_154458520 | 0.19 |
ENST00000486773.1
|
SHE
|
Src homology 2 domain containing E |
| chr1_-_236046872 | 0.18 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr18_+_32073253 | 0.18 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chrX_+_23801280 | 0.17 |
ENST00000379251.3
ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1
|
spermidine/spermine N1-acetyltransferase 1 |
| chr7_-_38403077 | 0.17 |
ENST00000426402.2
|
TRGV2
|
T cell receptor gamma variable 2 |
| chr17_-_39623681 | 0.17 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr7_+_16793160 | 0.17 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr11_-_130786400 | 0.16 |
ENST00000265909.4
|
SNX19
|
sorting nexin 19 |
| chr11_-_18548426 | 0.15 |
ENST00000357193.3
ENST00000536719.1 |
TSG101
|
tumor susceptibility 101 |
| chr10_-_71906342 | 0.15 |
ENST00000287078.6
ENST00000335494.5 |
TYSND1
|
trypsin domain containing 1 |
| chr20_+_62289640 | 0.15 |
ENST00000508582.2
ENST00000360203.5 ENST00000356810.4 |
RTEL1
|
regulator of telomere elongation helicase 1 |
| chr6_+_123100853 | 0.14 |
ENST00000356535.4
|
FABP7
|
fatty acid binding protein 7, brain |
| chr19_+_47852538 | 0.14 |
ENST00000328771.4
|
DHX34
|
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
| chr5_+_75379224 | 0.14 |
ENST00000322285.7
|
SV2C
|
synaptic vesicle glycoprotein 2C |
| chr7_+_100081542 | 0.14 |
ENST00000300179.2
ENST00000423930.1 |
NYAP1
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1 |
| chr19_-_55549624 | 0.13 |
ENST00000417454.1
ENST00000310373.3 ENST00000333884.2 |
GP6
|
glycoprotein VI (platelet) |
| chrX_-_154563889 | 0.13 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr2_-_136594740 | 0.12 |
ENST00000264162.2
|
LCT
|
lactase |
| chr11_-_82782952 | 0.12 |
ENST00000534141.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr17_-_56595196 | 0.11 |
ENST00000579921.1
ENST00000579925.1 ENST00000323456.5 |
MTMR4
|
myotubularin related protein 4 |
| chr7_-_38394118 | 0.10 |
ENST00000390345.2
|
TRGV4
|
T cell receptor gamma variable 4 |
| chr11_-_130786333 | 0.10 |
ENST00000533214.1
ENST00000528555.1 ENST00000530356.1 ENST00000539184.1 |
SNX19
|
sorting nexin 19 |
| chr17_+_72983674 | 0.09 |
ENST00000337231.5
|
CDR2L
|
cerebellar degeneration-related protein 2-like |
| chr4_-_140216948 | 0.09 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr5_-_169725231 | 0.08 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr4_+_159236462 | 0.07 |
ENST00000460056.2
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr4_-_5894777 | 0.06 |
ENST00000324989.7
|
CRMP1
|
collapsin response mediator protein 1 |
| chr1_+_159141397 | 0.06 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr17_-_10325261 | 0.06 |
ENST00000403437.2
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr1_+_154229547 | 0.06 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr6_+_12958137 | 0.06 |
ENST00000457702.2
ENST00000379345.2 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr2_-_25565377 | 0.05 |
ENST00000264709.3
ENST00000406659.3 |
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr1_-_111148241 | 0.05 |
ENST00000440270.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr4_-_17783135 | 0.05 |
ENST00000265018.3
|
FAM184B
|
family with sequence similarity 184, member B |
| chr13_+_27998681 | 0.04 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr3_-_149688502 | 0.03 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chr19_-_19006890 | 0.03 |
ENST00000247005.6
|
GDF1
|
growth differentiation factor 1 |
| chr3_+_98250743 | 0.03 |
ENST00000284311.3
|
GPR15
|
G protein-coupled receptor 15 |
| chr9_+_74526532 | 0.03 |
ENST00000486911.2
|
C9orf85
|
chromosome 9 open reading frame 85 |
| chr5_+_112074029 | 0.03 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr3_+_178276488 | 0.02 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr5_+_74807886 | 0.02 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chrX_+_70364667 | 0.02 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr11_+_67007518 | 0.02 |
ENST00000530342.1
ENST00000308783.5 |
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr6_+_123100620 | 0.02 |
ENST00000368444.3
|
FABP7
|
fatty acid binding protein 7, brain |
| chr3_-_195163803 | 0.02 |
ENST00000326793.6
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr14_+_60715928 | 0.01 |
ENST00000395076.4
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr1_-_155270770 | 0.01 |
ENST00000392414.3
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr3_-_149688655 | 0.01 |
ENST00000461930.1
ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2
|
profilin 2 |
| chr8_+_42196000 | 0.00 |
ENST00000518925.1
ENST00000538005.1 |
POLB
|
polymerase (DNA directed), beta |
| chr21_-_48024986 | 0.00 |
ENST00000291700.4
ENST00000367071.4 |
S100B
|
S100 calcium binding protein B |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.3 | 1.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 3.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.2 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.3 | GO:0072237 | distal tubule morphogenesis(GO:0072156) metanephric proximal tubule development(GO:0072237) |
| 0.1 | 0.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.3 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.5 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.1 | 0.2 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.2 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.5 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.5 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.6 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 1.0 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.4 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.4 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.4 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.0 | GO:0072678 | T cell migration(GO:0072678) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.4 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.5 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 3.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 3.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |