Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
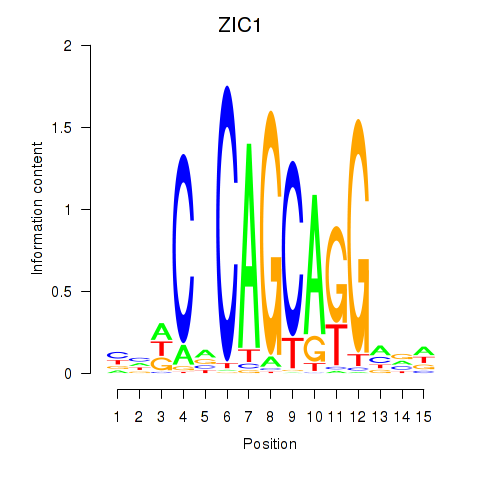
Results for ZIC1
Z-value: 0.71
Transcription factors associated with ZIC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC1
|
ENSG00000152977.5 | Zic family member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC1 | hg19_v2_chr3_+_147127142_147127171 | 0.02 | 9.3e-01 | Click! |
Activity profile of ZIC1 motif
Sorted Z-values of ZIC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_2182388 | 0.98 |
ENST00000421783.1
ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS
INS-IGF2
|
insulin INS-IGF2 readthrough |
| chr17_-_39203519 | 0.93 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr1_-_177134024 | 0.93 |
ENST00000367654.3
|
ASTN1
|
astrotactin 1 |
| chr6_-_35656685 | 0.92 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr3_-_58563094 | 0.87 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A |
| chr6_-_35656712 | 0.86 |
ENST00000357266.4
ENST00000542713.1 |
FKBP5
|
FK506 binding protein 5 |
| chr19_-_6433765 | 0.75 |
ENST00000321510.6
|
SLC25A41
|
solute carrier family 25, member 41 |
| chr16_-_755819 | 0.73 |
ENST00000397621.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr22_-_24622080 | 0.65 |
ENST00000425408.1
|
GGT5
|
gamma-glutamyltransferase 5 |
| chr1_-_60392452 | 0.63 |
ENST00000371204.3
|
CYP2J2
|
cytochrome P450, family 2, subfamily J, polypeptide 2 |
| chr2_+_54951679 | 0.50 |
ENST00000356458.6
|
EML6
|
echinoderm microtubule associated protein like 6 |
| chr2_+_120189422 | 0.46 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr10_+_118350468 | 0.45 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr13_-_29069232 | 0.44 |
ENST00000282397.4
ENST00000541932.1 ENST00000539099.1 |
FLT1
|
fms-related tyrosine kinase 1 |
| chr14_+_22739823 | 0.44 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr9_-_127263265 | 0.44 |
ENST00000373587.3
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr2_-_29297127 | 0.43 |
ENST00000331664.5
|
C2orf71
|
chromosome 2 open reading frame 71 |
| chr16_-_745946 | 0.43 |
ENST00000562563.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr19_-_8213753 | 0.42 |
ENST00000601739.1
|
FBN3
|
fibrillin 3 |
| chr19_+_41509851 | 0.41 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr21_+_46066331 | 0.40 |
ENST00000334670.8
|
KRTAP10-11
|
keratin associated protein 10-11 |
| chr17_-_39183452 | 0.40 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr1_+_16348366 | 0.40 |
ENST00000375692.1
ENST00000420078.1 |
CLCNKA
|
chloride channel, voltage-sensitive Ka |
| chr8_+_25042192 | 0.39 |
ENST00000410074.1
|
DOCK5
|
dedicator of cytokinesis 5 |
| chr4_-_7105056 | 0.38 |
ENST00000504402.1
ENST00000499242.2 ENST00000501888.2 |
RP11-367J11.3
|
RP11-367J11.3 |
| chr11_-_110582773 | 0.36 |
ENST00000524756.1
|
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr16_-_31100284 | 0.36 |
ENST00000280606.6
|
PRSS53
|
protease, serine, 53 |
| chr21_+_46020497 | 0.36 |
ENST00000380102.2
|
KRTAP10-7
|
keratin associated protein 10-7 |
| chr10_+_118350522 | 0.36 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr14_-_65409502 | 0.35 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr21_-_46012386 | 0.34 |
ENST00000400368.1
|
KRTAP10-6
|
keratin associated protein 10-6 |
| chr8_+_85097110 | 0.34 |
ENST00000517638.1
ENST00000522647.1 |
RALYL
|
RALY RNA binding protein-like |
| chr2_-_46769694 | 0.34 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr11_-_59612969 | 0.34 |
ENST00000541311.1
ENST00000257248.2 |
GIF
|
gastric intrinsic factor (vitamin B synthesis) |
| chr1_+_1567474 | 0.33 |
ENST00000356026.5
|
MMP23B
|
matrix metallopeptidase 23B |
| chr1_+_16370271 | 0.32 |
ENST00000375679.4
|
CLCNKB
|
chloride channel, voltage-sensitive Kb |
| chr1_+_1567546 | 0.32 |
ENST00000378675.3
|
MMP23B
|
matrix metallopeptidase 23B |
| chr17_-_39216344 | 0.31 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr7_-_37026108 | 0.31 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr20_+_31619454 | 0.31 |
ENST00000349552.1
|
BPIFB6
|
BPI fold containing family B, member 6 |
| chr19_+_13135731 | 0.31 |
ENST00000587260.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_+_16348497 | 0.30 |
ENST00000439316.2
|
CLCNKA
|
chloride channel, voltage-sensitive Ka |
| chr17_+_60704762 | 0.29 |
ENST00000303375.5
|
MRC2
|
mannose receptor, C type 2 |
| chr14_-_65409438 | 0.29 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr1_-_211307315 | 0.29 |
ENST00000271751.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr4_-_66535653 | 0.28 |
ENST00000354839.4
ENST00000432638.2 |
EPHA5
|
EPH receptor A5 |
| chr15_-_71407833 | 0.27 |
ENST00000449977.2
|
CT62
|
cancer/testis antigen 62 |
| chr1_-_1711508 | 0.27 |
ENST00000378625.1
|
NADK
|
NAD kinase |
| chr15_-_71407806 | 0.27 |
ENST00000566432.1
ENST00000567117.1 |
CT62
|
cancer/testis antigen 62 |
| chrX_+_71354000 | 0.26 |
ENST00000510661.1
ENST00000535692.1 |
NHSL2
|
NHS-like 2 |
| chr6_+_44187242 | 0.25 |
ENST00000393844.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr2_+_105050794 | 0.25 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr14_+_57735614 | 0.24 |
ENST00000261558.3
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr5_+_140514782 | 0.24 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr4_-_66536196 | 0.24 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr21_+_45148735 | 0.24 |
ENST00000327574.4
|
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr16_+_23194033 | 0.24 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chrX_-_129299847 | 0.23 |
ENST00000319908.3
ENST00000287295.3 |
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr9_-_16870704 | 0.23 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chrX_+_68835911 | 0.23 |
ENST00000525810.1
ENST00000527388.1 ENST00000374553.2 ENST00000374552.4 ENST00000338901.3 ENST00000524573.1 |
EDA
|
ectodysplasin A |
| chr19_+_45409011 | 0.22 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr21_+_46117087 | 0.22 |
ENST00000400365.3
|
KRTAP10-12
|
keratin associated protein 10-12 |
| chr2_-_74780176 | 0.22 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr17_-_39197699 | 0.22 |
ENST00000306271.4
|
KRTAP1-1
|
keratin associated protein 1-1 |
| chr4_-_109090106 | 0.21 |
ENST00000379951.2
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr1_-_244615425 | 0.21 |
ENST00000366535.3
|
ADSS
|
adenylosuccinate synthase |
| chrX_-_129299638 | 0.21 |
ENST00000535724.1
ENST00000346424.2 |
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr3_+_71803201 | 0.21 |
ENST00000304411.2
|
GPR27
|
G protein-coupled receptor 27 |
| chr19_+_13135790 | 0.21 |
ENST00000358552.3
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr7_+_150264365 | 0.20 |
ENST00000255945.2
ENST00000461940.1 |
GIMAP4
|
GTPase, IMAP family member 4 |
| chr4_-_109089573 | 0.20 |
ENST00000265165.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr13_+_102104952 | 0.20 |
ENST00000376180.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chrX_-_78622805 | 0.19 |
ENST00000373298.2
|
ITM2A
|
integral membrane protein 2A |
| chr1_+_207038699 | 0.18 |
ENST00000367098.1
|
IL20
|
interleukin 20 |
| chr10_-_81708854 | 0.18 |
ENST00000372292.3
|
SFTPD
|
surfactant protein D |
| chr2_-_208989225 | 0.18 |
ENST00000264376.4
|
CRYGD
|
crystallin, gamma D |
| chr4_-_175750364 | 0.18 |
ENST00000340217.5
ENST00000274093.3 |
GLRA3
|
glycine receptor, alpha 3 |
| chr17_-_3796334 | 0.17 |
ENST00000381771.2
ENST00000348335.2 ENST00000158166.5 |
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr21_+_45209394 | 0.17 |
ENST00000497547.1
|
RRP1
|
ribosomal RNA processing 1 |
| chr13_-_100624012 | 0.17 |
ENST00000267294.4
|
ZIC5
|
Zic family member 5 |
| chr1_+_207039154 | 0.17 |
ENST00000367096.3
ENST00000391930.2 |
IL20
|
interleukin 20 |
| chr19_-_2740036 | 0.17 |
ENST00000586572.1
ENST00000269740.4 |
AC006538.4
SLC39A3
|
Uncharacterized protein solute carrier family 39 (zinc transporter), member 3 |
| chrX_+_70364667 | 0.16 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr4_-_66536057 | 0.16 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr1_+_46668994 | 0.16 |
ENST00000371980.3
|
LURAP1
|
leucine rich adaptor protein 1 |
| chr6_-_34216766 | 0.16 |
ENST00000481533.1
ENST00000468145.1 ENST00000413013.2 ENST00000394990.4 ENST00000335352.3 |
C6orf1
|
chromosome 6 open reading frame 1 |
| chrX_+_24711997 | 0.16 |
ENST00000379068.3
ENST00000379059.3 |
POLA1
|
polymerase (DNA directed), alpha 1, catalytic subunit |
| chr1_-_111991908 | 0.16 |
ENST00000235090.5
|
WDR77
|
WD repeat domain 77 |
| chr18_+_76829441 | 0.15 |
ENST00000458297.2
|
ATP9B
|
ATPase, class II, type 9B |
| chr15_+_80351910 | 0.15 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr21_+_31768348 | 0.15 |
ENST00000355459.2
|
KRTAP13-1
|
keratin associated protein 13-1 |
| chr19_-_3028354 | 0.14 |
ENST00000586422.1
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr2_-_74779744 | 0.14 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr3_-_53080047 | 0.13 |
ENST00000482396.1
ENST00000358080.2 ENST00000296295.6 ENST00000394752.3 |
SFMBT1
|
Scm-like with four mbt domains 1 |
| chr10_+_49917751 | 0.13 |
ENST00000325239.5
ENST00000413659.2 |
WDFY4
|
WDFY family member 4 |
| chr11_-_57417405 | 0.13 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr21_-_39870339 | 0.13 |
ENST00000429727.2
ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr15_+_41245160 | 0.13 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr14_+_38677123 | 0.13 |
ENST00000267377.2
|
SSTR1
|
somatostatin receptor 1 |
| chr1_+_22963158 | 0.13 |
ENST00000438241.1
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr17_-_39191107 | 0.13 |
ENST00000344363.5
|
KRTAP1-3
|
keratin associated protein 1-3 |
| chr11_-_62689046 | 0.13 |
ENST00000306960.3
ENST00000543973.1 |
CHRM1
|
cholinergic receptor, muscarinic 1 |
| chr20_+_61924532 | 0.12 |
ENST00000358894.6
ENST00000326996.6 ENST00000435874.1 |
COL20A1
|
collagen, type XX, alpha 1 |
| chr19_+_35940486 | 0.12 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr15_+_73976715 | 0.12 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr17_-_3794021 | 0.12 |
ENST00000381769.2
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr8_-_120685608 | 0.12 |
ENST00000427067.2
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr18_-_52989217 | 0.12 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr1_+_18958008 | 0.12 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr9_-_35103105 | 0.12 |
ENST00000452248.2
ENST00000356493.5 |
STOML2
|
stomatin (EPB72)-like 2 |
| chr3_-_45838011 | 0.12 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr12_-_11002063 | 0.12 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr21_-_31864275 | 0.12 |
ENST00000334063.4
|
KRTAP19-3
|
keratin associated protein 19-3 |
| chr16_+_838614 | 0.12 |
ENST00000262315.9
ENST00000455171.2 |
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
| chr11_-_46142505 | 0.11 |
ENST00000524497.1
ENST00000418153.2 |
PHF21A
|
PHD finger protein 21A |
| chr19_-_14586125 | 0.11 |
ENST00000292513.3
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa |
| chr6_-_117086873 | 0.11 |
ENST00000368557.4
|
FAM162B
|
family with sequence similarity 162, member B |
| chr3_+_14444063 | 0.11 |
ENST00000454876.2
ENST00000360861.3 ENST00000416216.2 |
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr2_+_90248739 | 0.11 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr17_-_43045439 | 0.11 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr16_-_11485922 | 0.11 |
ENST00000599216.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr22_-_23922448 | 0.11 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr3_+_126243126 | 0.10 |
ENST00000319340.2
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13 |
| chr11_+_66824346 | 0.10 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr2_+_89184868 | 0.10 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr22_-_23922410 | 0.10 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr21_+_18811205 | 0.09 |
ENST00000440664.1
|
C21orf37
|
chromosome 21 open reading frame 37 |
| chrX_+_53078273 | 0.09 |
ENST00000332582.4
|
GPR173
|
G protein-coupled receptor 173 |
| chr22_+_21369316 | 0.09 |
ENST00000413302.2
ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chrX_+_72064876 | 0.09 |
ENST00000373532.3
ENST00000333826.5 |
DMRTC1B
|
DMRT-like family C1B |
| chr18_+_76829385 | 0.09 |
ENST00000426216.2
ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B
|
ATPase, class II, type 9B |
| chr1_+_22962948 | 0.09 |
ENST00000374642.3
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr7_+_39017504 | 0.09 |
ENST00000403058.1
|
POU6F2
|
POU class 6 homeobox 2 |
| chr22_+_29702572 | 0.09 |
ENST00000407647.2
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr1_-_71513471 | 0.09 |
ENST00000370931.3
ENST00000356595.4 ENST00000306666.5 ENST00000370932.2 ENST00000351052.5 ENST00000414819.1 ENST00000370924.4 |
PTGER3
|
prostaglandin E receptor 3 (subtype EP3) |
| chr11_+_67777751 | 0.09 |
ENST00000316367.6
ENST00000007633.8 ENST00000342456.6 |
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chrX_-_153979315 | 0.09 |
ENST00000369575.3
ENST00000369568.4 ENST00000424127.2 |
GAB3
|
GRB2-associated binding protein 3 |
| chr13_+_30510003 | 0.09 |
ENST00000400540.1
|
LINC00544
|
long intergenic non-protein coding RNA 544 |
| chr4_-_89444883 | 0.09 |
ENST00000273968.4
|
PYURF
|
PIGY upstream reading frame |
| chr4_-_47839966 | 0.09 |
ENST00000273857.4
ENST00000505909.1 ENST00000502252.1 |
CORIN
|
corin, serine peptidase |
| chr12_-_27091183 | 0.09 |
ENST00000544548.1
ENST00000261191.7 ENST00000537336.1 |
ASUN
|
asunder spermatogenesis regulator |
| chr1_+_61869748 | 0.08 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr3_-_45837959 | 0.08 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr7_-_4877677 | 0.08 |
ENST00000538469.1
|
RADIL
|
Ras association and DIL domains |
| chrX_+_72064690 | 0.08 |
ENST00000438696.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr14_-_50999190 | 0.07 |
ENST00000557390.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr19_+_54024251 | 0.07 |
ENST00000253144.9
|
ZNF331
|
zinc finger protein 331 |
| chr2_+_234668894 | 0.07 |
ENST00000305208.5
ENST00000608383.1 ENST00000360418.3 |
UGT1A8
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr2_-_98280383 | 0.07 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr15_+_81589254 | 0.07 |
ENST00000394652.2
|
IL16
|
interleukin 16 |
| chrX_+_13707235 | 0.07 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr9_-_112260531 | 0.06 |
ENST00000374541.2
ENST00000262539.3 |
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr6_+_41755389 | 0.06 |
ENST00000398884.3
ENST00000398881.3 |
TOMM6
|
translocase of outer mitochondrial membrane 6 homolog (yeast) |
| chr3_-_79816965 | 0.06 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr19_-_2739992 | 0.06 |
ENST00000545664.1
ENST00000589363.1 ENST00000455372.2 |
SLC39A3
|
solute carrier family 39 (zinc transporter), member 3 |
| chr5_+_167181917 | 0.06 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr4_-_40632605 | 0.06 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr10_-_36813162 | 0.06 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr9_+_35792151 | 0.06 |
ENST00000342694.2
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr14_+_23340822 | 0.06 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr9_+_132934835 | 0.05 |
ENST00000372398.3
|
NCS1
|
neuronal calcium sensor 1 |
| chr17_-_53809473 | 0.05 |
ENST00000575734.1
|
TMEM100
|
transmembrane protein 100 |
| chr6_+_31540056 | 0.05 |
ENST00000418386.2
|
LTA
|
lymphotoxin alpha |
| chr6_+_31949801 | 0.05 |
ENST00000428956.2
ENST00000498271.1 |
C4A
|
complement component 4A (Rodgers blood group) |
| chr2_+_46769798 | 0.05 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr6_-_168476511 | 0.04 |
ENST00000440994.2
|
FRMD1
|
FERM domain containing 1 |
| chrY_+_4868267 | 0.04 |
ENST00000333703.4
|
PCDH11Y
|
protocadherin 11 Y-linked |
| chr3_+_141105705 | 0.04 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr4_-_121993673 | 0.04 |
ENST00000379692.4
|
NDNF
|
neuron-derived neurotrophic factor |
| chr9_+_124329336 | 0.04 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr17_-_76274572 | 0.04 |
ENST00000374945.1
|
RP11-219G17.4
|
RP11-219G17.4 |
| chr3_-_50329835 | 0.04 |
ENST00000429673.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr19_-_16582754 | 0.04 |
ENST00000602151.1
ENST00000597937.1 ENST00000535753.2 |
EPS15L1
|
epidermal growth factor receptor pathway substrate 15-like 1 |
| chr5_-_126409159 | 0.04 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr17_+_18280976 | 0.04 |
ENST00000399134.4
|
EVPLL
|
envoplakin-like |
| chrX_+_52926322 | 0.04 |
ENST00000430150.2
ENST00000452021.2 ENST00000412319.1 |
FAM156B
|
family with sequence similarity 156, member B |
| chr22_+_41956767 | 0.04 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr3_+_32859510 | 0.04 |
ENST00000383763.5
|
TRIM71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr19_+_10563567 | 0.04 |
ENST00000344979.3
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr10_-_101945771 | 0.04 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr12_-_15038779 | 0.03 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr16_+_28858004 | 0.03 |
ENST00000322610.8
|
SH2B1
|
SH2B adaptor protein 1 |
| chr1_-_111991850 | 0.03 |
ENST00000411751.2
|
WDR77
|
WD repeat domain 77 |
| chr7_-_102789629 | 0.03 |
ENST00000417955.1
ENST00000341533.4 ENST00000425379.1 |
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr15_+_85523671 | 0.03 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr1_+_111992064 | 0.03 |
ENST00000483994.1
|
ATP5F1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 |
| chr10_-_49701686 | 0.03 |
ENST00000417247.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr5_-_76935513 | 0.03 |
ENST00000306422.3
|
OTP
|
orthopedia homeobox |
| chr22_+_44319648 | 0.03 |
ENST00000423180.2
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr22_+_44319619 | 0.03 |
ENST00000216180.3
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr15_+_77712993 | 0.03 |
ENST00000336216.4
ENST00000381714.3 ENST00000558651.1 |
HMG20A
|
high mobility group 20A |
| chr1_+_89990431 | 0.03 |
ENST00000330947.2
ENST00000358200.4 |
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr2_-_27938593 | 0.02 |
ENST00000379677.2
|
AC074091.13
|
Uncharacterized protein |
| chr11_-_74022658 | 0.02 |
ENST00000427714.2
ENST00000331597.4 |
P4HA3
|
prolyl 4-hydroxylase, alpha polypeptide III |
| chr7_-_107880508 | 0.02 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr7_+_29874341 | 0.02 |
ENST00000409290.1
ENST00000242140.5 |
WIPF3
|
WAS/WASL interacting protein family, member 3 |
| chr17_-_18945798 | 0.02 |
ENST00000395635.1
|
GRAP
|
GRB2-related adaptor protein |
| chr1_-_89641680 | 0.02 |
ENST00000294671.2
|
GBP7
|
guanylate binding protein 7 |
| chr2_-_89399845 | 0.02 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr2_+_169926047 | 0.02 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.1 | 0.6 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.4 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.4 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.2 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.1 | 0.2 | GO:2000646 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.4 | GO:1904044 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) response to aldosterone(GO:1904044) |
| 0.1 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.8 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 1.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.8 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 1.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 1.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.7 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.4 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 1.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.1 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.4 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 3.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.2 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 2.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.1 | 0.2 | GO:0008478 | pyridoxal kinase activity(GO:0008478) lithium ion binding(GO:0031403) |
| 0.1 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.2 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 1.0 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.8 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.4 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |