Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
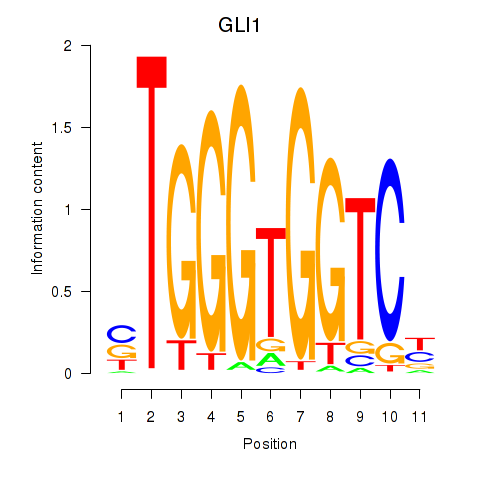
Results for ZIC2_GLI1
Z-value: 0.71
Transcription factors associated with ZIC2_GLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC2
|
ENSG00000043355.6 | Zic family member 2 |
|
GLI1
|
ENSG00000111087.5 | GLI family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI1 | hg19_v2_chr12_+_57853918_57853934, hg19_v2_chr12_+_57857475_57857475 | -0.36 | 5.0e-02 | Click! |
| ZIC2 | hg19_v2_chr13_+_100634004_100634026 | -0.04 | 8.4e-01 | Click! |
Activity profile of ZIC2_GLI1 motif
Sorted Z-values of ZIC2_GLI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_163148973 | 4.15 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chr9_-_34397800 | 3.34 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr22_-_50970506 | 2.31 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chrX_+_152240819 | 2.09 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr9_+_131218698 | 1.83 |
ENST00000434106.3
ENST00000546203.1 ENST00000446274.1 ENST00000421776.2 ENST00000432065.2 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr22_-_50970566 | 1.74 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr1_+_47264711 | 1.65 |
ENST00000371923.4
ENST00000271153.4 ENST00000371919.4 |
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr6_-_29595779 | 1.59 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr15_-_93616892 | 1.53 |
ENST00000556658.1
ENST00000538818.1 ENST00000425933.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr22_-_50970919 | 1.47 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr9_+_131219179 | 1.46 |
ENST00000372791.3
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chr9_+_131218408 | 1.45 |
ENST00000351030.3
ENST00000604420.1 ENST00000535026.1 ENST00000448249.3 ENST00000393527.3 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr16_+_75256507 | 1.34 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1 |
| chr15_-_78526855 | 1.34 |
ENST00000541759.1
ENST00000558130.1 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr2_-_28113965 | 1.32 |
ENST00000302188.3
|
RBKS
|
ribokinase |
| chr9_+_131218336 | 1.14 |
ENST00000372814.3
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chr12_-_25801478 | 1.08 |
ENST00000540106.1
ENST00000445693.1 ENST00000545543.1 ENST00000542224.1 |
IFLTD1
|
intermediate filament tail domain containing 1 |
| chr20_+_57430162 | 0.95 |
ENST00000450130.1
ENST00000349036.3 ENST00000423897.1 |
GNAS
|
GNAS complex locus |
| chr3_-_52486841 | 0.90 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr11_-_75379612 | 0.86 |
ENST00000526740.1
|
MAP6
|
microtubule-associated protein 6 |
| chr17_+_17876127 | 0.70 |
ENST00000582416.1
ENST00000313838.8 ENST00000411504.2 ENST00000581264.1 ENST00000399187.1 ENST00000479684.2 ENST00000584166.1 ENST00000585108.1 ENST00000399182.1 ENST00000579977.1 |
LRRC48
|
leucine rich repeat containing 48 |
| chr11_-_111783919 | 0.67 |
ENST00000531198.1
ENST00000533879.1 |
CRYAB
|
crystallin, alpha B |
| chr3_+_113616317 | 0.67 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr2_-_213403565 | 0.66 |
ENST00000342788.4
ENST00000436443.1 |
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
| chr1_-_217250231 | 0.64 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr2_+_149402553 | 0.63 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr16_-_16317321 | 0.63 |
ENST00000205557.7
ENST00000575728.1 |
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chr7_+_69064300 | 0.62 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr2_+_97481974 | 0.60 |
ENST00000377060.3
ENST00000305510.3 |
CNNM3
|
cyclin M3 |
| chr7_-_112726393 | 0.60 |
ENST00000449591.1
ENST00000449735.1 ENST00000438062.1 ENST00000424100.1 |
GPR85
|
G protein-coupled receptor 85 |
| chr17_+_7608511 | 0.58 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr7_+_48211048 | 0.56 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr3_+_49449636 | 0.52 |
ENST00000273590.3
|
TCTA
|
T-cell leukemia translocation altered |
| chr12_-_122985067 | 0.52 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr12_-_45270151 | 0.51 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr17_+_40834580 | 0.51 |
ENST00000264638.4
|
CNTNAP1
|
contactin associated protein 1 |
| chr6_-_55739542 | 0.49 |
ENST00000446683.2
|
BMP5
|
bone morphogenetic protein 5 |
| chr17_-_26989136 | 0.49 |
ENST00000247020.4
|
SDF2
|
stromal cell-derived factor 2 |
| chr16_+_2521500 | 0.49 |
ENST00000293973.1
|
NTN3
|
netrin 3 |
| chr12_-_122985494 | 0.49 |
ENST00000336229.4
|
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr16_-_1031259 | 0.47 |
ENST00000563837.1
ENST00000563863.1 ENST00000565069.1 ENST00000570014.1 |
RP11-161M6.2
LMF1
|
RP11-161M6.2 lipase maturation factor 1 |
| chr4_-_5021164 | 0.45 |
ENST00000506508.1
ENST00000509419.1 ENST00000307746.4 |
CYTL1
|
cytokine-like 1 |
| chr16_+_88493879 | 0.45 |
ENST00000565624.1
ENST00000437464.1 |
ZNF469
|
zinc finger protein 469 |
| chr14_-_61124977 | 0.44 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr6_+_33172407 | 0.44 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr15_-_78526942 | 0.43 |
ENST00000258873.4
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr12_-_45270077 | 0.43 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr11_+_45868957 | 0.40 |
ENST00000443527.2
|
CRY2
|
cryptochrome 2 (photolyase-like) |
| chr17_-_74533734 | 0.40 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr1_+_32674675 | 0.39 |
ENST00000409358.1
|
DCDC2B
|
doublecortin domain containing 2B |
| chr10_-_95360983 | 0.38 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr8_-_17555164 | 0.38 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr14_+_24583836 | 0.38 |
ENST00000559115.1
ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr2_-_46769694 | 0.38 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr14_-_64971288 | 0.36 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr16_+_30675654 | 0.36 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr7_-_105517021 | 0.35 |
ENST00000318724.4
ENST00000419735.3 |
ATXN7L1
|
ataxin 7-like 1 |
| chr12_-_6798616 | 0.35 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr9_-_73483958 | 0.34 |
ENST00000377101.1
ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr12_-_6798410 | 0.33 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr7_+_119913688 | 0.33 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr1_-_151798546 | 0.33 |
ENST00000356728.6
|
RORC
|
RAR-related orphan receptor C |
| chr12_-_6798523 | 0.33 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr7_-_99679324 | 0.32 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr14_+_24584508 | 0.32 |
ENST00000559354.1
ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr7_+_116312411 | 0.31 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr1_-_112046289 | 0.31 |
ENST00000241356.4
|
ADORA3
|
adenosine A3 receptor |
| chr1_+_36690011 | 0.30 |
ENST00000354618.5
ENST00000469141.2 ENST00000478853.1 |
THRAP3
|
thyroid hormone receptor associated protein 3 |
| chr17_-_74533963 | 0.30 |
ENST00000293230.5
|
CYGB
|
cytoglobin |
| chr20_-_30310656 | 0.29 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr15_+_60296421 | 0.29 |
ENST00000396057.4
|
FOXB1
|
forkhead box B1 |
| chr3_+_4535025 | 0.28 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr1_+_152943122 | 0.28 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr9_+_22446814 | 0.28 |
ENST00000325870.2
|
DMRTA1
|
DMRT-like family A1 |
| chr6_+_31620191 | 0.28 |
ENST00000375918.2
ENST00000375920.4 |
APOM
|
apolipoprotein M |
| chr6_+_26240561 | 0.28 |
ENST00000377745.2
|
HIST1H4F
|
histone cluster 1, H4f |
| chr17_+_43239231 | 0.27 |
ENST00000591576.1
ENST00000591070.1 ENST00000592695.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr20_+_36149602 | 0.27 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr9_-_130639997 | 0.27 |
ENST00000373176.1
|
AK1
|
adenylate kinase 1 |
| chr4_-_186697044 | 0.27 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_+_153839149 | 0.26 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr3_-_8686479 | 0.26 |
ENST00000544814.1
ENST00000427408.1 |
SSUH2
|
ssu-2 homolog (C. elegans) |
| chr7_+_149412094 | 0.26 |
ENST00000255992.10
ENST00000319551.8 |
KRBA1
|
KRAB-A domain containing 1 |
| chr2_+_217498105 | 0.26 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr1_+_162531294 | 0.25 |
ENST00000367926.4
ENST00000271469.3 |
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1 |
| chr17_-_15496722 | 0.25 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr1_-_205782304 | 0.25 |
ENST00000367137.3
|
SLC41A1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr7_+_31009148 | 0.25 |
ENST00000409904.3
ENST00000409316.1 |
GHRHR
|
growth hormone releasing hormone receptor |
| chr3_+_4535155 | 0.25 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr20_-_30310693 | 0.25 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chr11_-_111784005 | 0.25 |
ENST00000527899.1
|
CRYAB
|
crystallin, alpha B |
| chr2_+_178977143 | 0.24 |
ENST00000286070.5
|
RBM45
|
RNA binding motif protein 45 |
| chr1_+_164528866 | 0.24 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr2_-_133427767 | 0.23 |
ENST00000397463.2
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr4_-_186696425 | 0.23 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr20_+_32782375 | 0.23 |
ENST00000568305.1
|
ASIP
|
agouti signaling protein |
| chr5_+_102455968 | 0.22 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr7_+_138145145 | 0.21 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr5_+_52776449 | 0.21 |
ENST00000396947.3
|
FST
|
follistatin |
| chr6_-_43337180 | 0.21 |
ENST00000318149.3
ENST00000361428.2 |
ZNF318
|
zinc finger protein 318 |
| chr6_-_134373732 | 0.21 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr2_+_26915584 | 0.21 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr7_-_105516923 | 0.21 |
ENST00000478915.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr21_-_27945562 | 0.21 |
ENST00000299340.4
ENST00000435845.2 |
CYYR1
|
cysteine/tyrosine-rich 1 |
| chr19_+_4198072 | 0.20 |
ENST00000262970.5
|
ANKRD24
|
ankyrin repeat domain 24 |
| chr9_-_86571628 | 0.20 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr22_-_30783356 | 0.20 |
ENST00000382363.3
|
RNF215
|
ring finger protein 215 |
| chr5_-_44388899 | 0.20 |
ENST00000264664.4
|
FGF10
|
fibroblast growth factor 10 |
| chr11_+_10477733 | 0.20 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr3_+_133118839 | 0.19 |
ENST00000302334.2
|
BFSP2
|
beaded filament structural protein 2, phakinin |
| chr7_-_91875109 | 0.19 |
ENST00000412043.2
ENST00000430102.1 ENST00000425073.1 ENST00000394503.2 ENST00000454017.1 ENST00000440209.1 ENST00000413688.1 ENST00000452773.1 ENST00000433016.1 ENST00000394505.2 ENST00000422347.1 ENST00000458493.1 ENST00000425919.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr22_-_46373004 | 0.19 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr11_-_111782696 | 0.18 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr6_-_29600832 | 0.18 |
ENST00000377016.4
ENST00000376977.3 ENST00000377034.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr1_-_183387723 | 0.18 |
ENST00000287713.6
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr22_-_30783075 | 0.18 |
ENST00000215798.6
|
RNF215
|
ring finger protein 215 |
| chr16_+_29819096 | 0.17 |
ENST00000568411.1
ENST00000563012.1 ENST00000562557.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr10_+_24755416 | 0.17 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr11_-_11643540 | 0.17 |
ENST00000227756.4
|
GALNT18
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18 |
| chr19_+_45312347 | 0.16 |
ENST00000270233.6
ENST00000591520.1 |
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr19_+_36208877 | 0.16 |
ENST00000420124.1
ENST00000222270.7 ENST00000341701.1 |
KMT2B
|
Histone-lysine N-methyltransferase 2B |
| chr2_+_137523086 | 0.16 |
ENST00000409968.1
|
THSD7B
|
thrombospondin, type I, domain containing 7B |
| chr19_-_55881741 | 0.15 |
ENST00000264563.2
ENST00000590625.1 ENST00000585513.1 |
IL11
|
interleukin 11 |
| chr2_+_106361333 | 0.15 |
ENST00000233154.4
ENST00000451463.2 |
NCK2
|
NCK adaptor protein 2 |
| chr5_+_71403061 | 0.15 |
ENST00000512974.1
ENST00000296755.7 |
MAP1B
|
microtubule-associated protein 1B |
| chr12_+_117176113 | 0.15 |
ENST00000319176.7
|
RNFT2
|
ring finger protein, transmembrane 2 |
| chr5_-_76935513 | 0.15 |
ENST00000306422.3
|
OTP
|
orthopedia homeobox |
| chrX_+_70443050 | 0.15 |
ENST00000361726.6
|
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr11_-_30607819 | 0.15 |
ENST00000448418.2
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr7_+_114562616 | 0.15 |
ENST00000448022.1
|
MDFIC
|
MyoD family inhibitor domain containing |
| chr12_+_122516626 | 0.15 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr17_+_6926339 | 0.15 |
ENST00000293805.5
|
BCL6B
|
B-cell CLL/lymphoma 6, member B |
| chr5_+_52776228 | 0.15 |
ENST00000256759.3
|
FST
|
follistatin |
| chr19_+_3708376 | 0.14 |
ENST00000539908.2
|
TJP3
|
tight junction protein 3 |
| chr9_-_98279241 | 0.14 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr20_-_34025999 | 0.14 |
ENST00000374369.3
|
GDF5
|
growth differentiation factor 5 |
| chr11_-_34938039 | 0.14 |
ENST00000395787.3
|
APIP
|
APAF1 interacting protein |
| chr14_+_96968802 | 0.14 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr1_+_226736446 | 0.14 |
ENST00000366788.3
ENST00000366789.4 |
C1orf95
|
chromosome 1 open reading frame 95 |
| chr1_+_154966058 | 0.14 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr11_-_30608413 | 0.14 |
ENST00000528686.1
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr17_-_7137582 | 0.14 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr16_+_29818857 | 0.13 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr5_+_67088974 | 0.13 |
ENST00000513234.1
ENST00000504068.1 ENST00000515227.1 |
RP11-434D9.1
|
RP11-434D9.1 |
| chr11_-_111783595 | 0.13 |
ENST00000528628.1
|
CRYAB
|
crystallin, alpha B |
| chr11_-_64527425 | 0.13 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr17_-_42298201 | 0.13 |
ENST00000527034.1
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr4_+_72052964 | 0.12 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr1_+_42846443 | 0.12 |
ENST00000410070.2
ENST00000431473.3 |
RIMKLA
|
ribosomal modification protein rimK-like family member A |
| chrX_-_107334790 | 0.12 |
ENST00000217958.3
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr16_+_29819372 | 0.12 |
ENST00000568544.1
ENST00000569978.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr19_+_4969116 | 0.12 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
| chr12_-_57522813 | 0.12 |
ENST00000556155.1
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr17_+_73997419 | 0.12 |
ENST00000425876.2
|
CDK3
|
cyclin-dependent kinase 3 |
| chr9_+_109625378 | 0.11 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr20_+_43343886 | 0.11 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr16_+_29819446 | 0.11 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr12_+_31477250 | 0.11 |
ENST00000313737.4
|
AC024940.1
|
AC024940.1 |
| chr1_-_150208320 | 0.11 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr9_-_115818951 | 0.11 |
ENST00000553380.1
ENST00000374227.3 |
ZFP37
|
ZFP37 zinc finger protein |
| chr11_-_119991589 | 0.11 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29 |
| chr2_-_178257401 | 0.11 |
ENST00000464747.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr4_-_38666430 | 0.11 |
ENST00000436901.1
|
AC021860.1
|
Uncharacterized protein |
| chr13_+_111767650 | 0.11 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr9_-_98079965 | 0.10 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C |
| chr4_-_134070250 | 0.10 |
ENST00000505289.1
ENST00000509715.1 |
RP11-9G1.3
|
RP11-9G1.3 |
| chr5_+_161275320 | 0.10 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr1_-_40105617 | 0.10 |
ENST00000372852.3
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr15_-_75743915 | 0.10 |
ENST00000394949.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr3_+_6902794 | 0.10 |
ENST00000357716.4
ENST00000486284.1 ENST00000389336.4 ENST00000403881.1 ENST00000402647.2 |
GRM7
|
glutamate receptor, metabotropic 7 |
| chr11_-_34937858 | 0.10 |
ENST00000278359.5
|
APIP
|
APAF1 interacting protein |
| chr6_-_33160231 | 0.09 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr11_+_64009072 | 0.09 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr2_+_159825143 | 0.09 |
ENST00000454300.1
ENST00000263635.6 |
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr11_-_111782484 | 0.09 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr12_-_49333446 | 0.09 |
ENST00000537495.1
|
AC073610.5
|
Uncharacterized protein |
| chr11_-_46142615 | 0.09 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr7_-_150777874 | 0.09 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr10_+_99344071 | 0.08 |
ENST00000370647.4
ENST00000370646.4 |
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr8_+_75736761 | 0.08 |
ENST00000260113.2
|
PI15
|
peptidase inhibitor 15 |
| chr20_+_43343517 | 0.08 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chrX_-_107334750 | 0.08 |
ENST00000340200.5
ENST00000372296.1 ENST00000372295.1 ENST00000361815.5 |
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr2_-_193059634 | 0.08 |
ENST00000392314.1
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr1_-_115053781 | 0.08 |
ENST00000358465.2
ENST00000369543.2 |
TRIM33
|
tripartite motif containing 33 |
| chr1_+_44679159 | 0.07 |
ENST00000315913.5
ENST00000372289.2 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr6_+_43968306 | 0.07 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr11_+_33278811 | 0.07 |
ENST00000303296.4
ENST00000379016.3 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr16_-_68002456 | 0.07 |
ENST00000576616.1
ENST00000572037.1 ENST00000338335.3 ENST00000422611.2 ENST00000316341.3 |
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr1_+_44679113 | 0.07 |
ENST00000361745.6
ENST00000446292.1 ENST00000440641.1 ENST00000436069.1 ENST00000437511.1 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr8_-_22014339 | 0.07 |
ENST00000306317.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr14_-_30396802 | 0.07 |
ENST00000415220.2
|
PRKD1
|
protein kinase D1 |
| chr10_-_86001210 | 0.07 |
ENST00000372105.3
|
LRIT1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr15_+_81591757 | 0.07 |
ENST00000558332.1
|
IL16
|
interleukin 16 |
| chr14_-_70655684 | 0.07 |
ENST00000356921.2
ENST00000381269.2 ENST00000357887.3 |
SLC8A3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr5_-_168727786 | 0.07 |
ENST00000332966.8
|
SLIT3
|
slit homolog 3 (Drosophila) |
| chrX_-_107979616 | 0.07 |
ENST00000372129.2
|
IRS4
|
insulin receptor substrate 4 |
| chr2_-_25016251 | 0.07 |
ENST00000328379.5
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr19_+_36602104 | 0.07 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
| chr12_-_4554780 | 0.06 |
ENST00000228837.2
|
FGF6
|
fibroblast growth factor 6 |
| chr6_-_32784687 | 0.06 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC2_GLI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.2 | 1.7 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.2 | 0.9 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.7 | GO:2000364 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.2 | 0.6 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.2 | 0.5 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.1 | 0.4 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.9 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.7 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 0.4 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.6 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.5 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 1.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 1.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.3 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 0.9 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.2 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.2 | GO:0072061 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) inner medullary collecting duct development(GO:0072061) |
| 0.1 | 0.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 10.0 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 1.8 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.5 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.1 | 0.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 1.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.3 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.8 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.2 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:1901675 | response to methylglyoxal(GO:0051595) negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0070269 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) pyroptosis(GO:0070269) |
| 0.0 | 0.4 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.6 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0002584 | negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.4 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.0 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.4 | 5.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 4.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 0.9 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 1.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 0.7 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 4.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 1.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.4 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.9 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.9 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 1.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.2 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.4 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.4 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |