Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for ZIC3_ZIC4
Z-value: 0.54
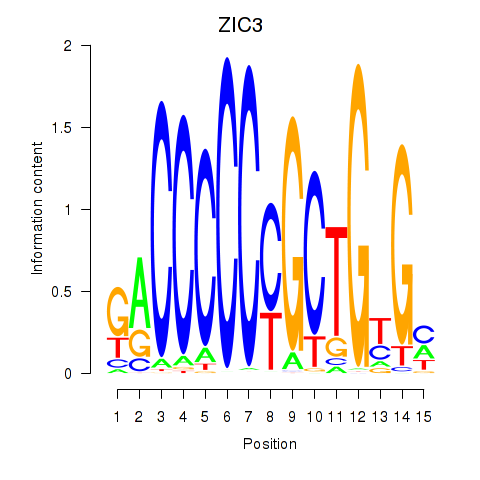
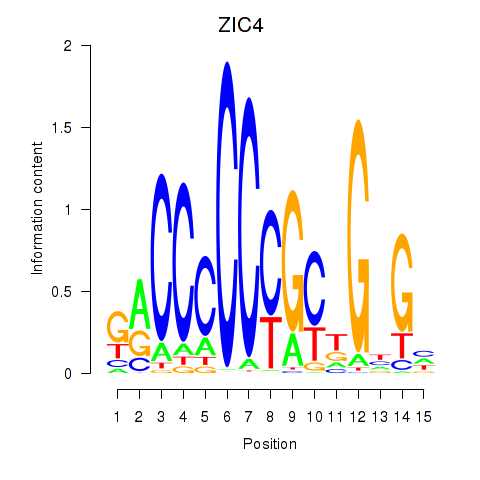
Transcription factors associated with ZIC3_ZIC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC3
|
ENSG00000156925.7 | Zic family member 3 |
|
ZIC4
|
ENSG00000174963.13 | Zic family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC4 | hg19_v2_chr3_-_147124547_147124647 | 0.37 | 4.2e-02 | Click! |
| ZIC3 | hg19_v2_chrX_+_136648297_136648319 | 0.13 | 4.9e-01 | Click! |
Activity profile of ZIC3_ZIC4 motif
Sorted Z-values of ZIC3_ZIC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_76506706 | 0.97 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr1_-_32801825 | 0.73 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr5_-_180018540 | 0.66 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr20_+_35201857 | 0.61 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr9_-_124989804 | 0.58 |
ENST00000373755.2
ENST00000373754.2 |
LHX6
|
LIM homeobox 6 |
| chr4_+_81951957 | 0.57 |
ENST00000282701.2
|
BMP3
|
bone morphogenetic protein 3 |
| chr2_-_99552620 | 0.57 |
ENST00000428096.1
ENST00000397899.2 ENST00000420294.1 |
KIAA1211L
|
KIAA1211-like |
| chr2_+_219187871 | 0.56 |
ENST00000258362.3
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr13_+_35516390 | 0.54 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr4_-_149363662 | 0.54 |
ENST00000355292.3
ENST00000358102.3 |
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chrX_-_34675391 | 0.52 |
ENST00000275954.3
|
TMEM47
|
transmembrane protein 47 |
| chr2_+_8822113 | 0.52 |
ENST00000396290.1
ENST00000331129.3 |
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr4_-_168155730 | 0.52 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr17_-_74137374 | 0.51 |
ENST00000322957.6
|
FOXJ1
|
forkhead box J1 |
| chr11_-_2182388 | 0.50 |
ENST00000421783.1
ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS
INS-IGF2
|
insulin INS-IGF2 readthrough |
| chr7_+_302918 | 0.49 |
ENST00000599994.1
|
AC187652.1
|
Protein LOC100996433 |
| chr20_+_35201993 | 0.48 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr11_-_67272794 | 0.48 |
ENST00000436757.2
ENST00000356404.3 |
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr2_+_238768187 | 0.45 |
ENST00000254661.4
ENST00000409726.1 |
RAMP1
|
receptor (G protein-coupled) activity modifying protein 1 |
| chr14_+_100848311 | 0.44 |
ENST00000542471.2
|
WDR25
|
WD repeat domain 25 |
| chr4_-_168155700 | 0.43 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr17_+_56160768 | 0.43 |
ENST00000579991.2
|
DYNLL2
|
dynein, light chain, LC8-type 2 |
| chr2_+_10183651 | 0.43 |
ENST00000305883.1
|
KLF11
|
Kruppel-like factor 11 |
| chr4_-_168155300 | 0.42 |
ENST00000541637.1
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_-_168155417 | 0.42 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr17_-_78194147 | 0.42 |
ENST00000534910.1
ENST00000326317.6 |
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr10_+_50818343 | 0.42 |
ENST00000374115.3
|
SLC18A3
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3 |
| chr17_-_5026397 | 0.41 |
ENST00000250076.3
|
ZNF232
|
zinc finger protein 232 |
| chr17_+_78194205 | 0.41 |
ENST00000573809.1
ENST00000361193.3 ENST00000574967.1 ENST00000576126.1 ENST00000411502.3 ENST00000546047.2 |
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr6_+_33168637 | 0.41 |
ENST00000374677.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr6_-_29595779 | 0.41 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr9_-_117880477 | 0.40 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr6_+_33168597 | 0.40 |
ENST00000374675.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr16_-_88717482 | 0.39 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr14_-_21994525 | 0.37 |
ENST00000538754.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr4_-_168155169 | 0.36 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr11_+_73676281 | 0.35 |
ENST00000543947.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr9_+_116638562 | 0.35 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr4_-_168155577 | 0.34 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr11_+_2466218 | 0.34 |
ENST00000155840.5
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr12_+_49932886 | 0.32 |
ENST00000257981.6
|
KCNH3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr12_+_56075330 | 0.32 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr12_-_51420128 | 0.30 |
ENST00000262051.7
ENST00000547732.1 ENST00000262052.5 ENST00000546488.1 ENST00000550714.1 ENST00000548193.1 ENST00000547579.1 ENST00000546743.1 |
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr21_+_46117087 | 0.30 |
ENST00000400365.3
|
KRTAP10-12
|
keratin associated protein 10-12 |
| chr12_-_51420108 | 0.29 |
ENST00000547198.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr10_+_28966271 | 0.29 |
ENST00000375533.3
|
BAMBI
|
BMP and activin membrane-bound inhibitor |
| chr11_+_73675873 | 0.29 |
ENST00000537753.1
ENST00000542350.1 |
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr12_-_51419924 | 0.29 |
ENST00000541174.2
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr12_+_51785057 | 0.27 |
ENST00000535225.2
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr1_-_38273840 | 0.26 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr9_-_140082983 | 0.26 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr22_-_47134077 | 0.25 |
ENST00000541677.1
ENST00000216264.8 |
CERK
|
ceramide kinase |
| chr1_+_3607228 | 0.25 |
ENST00000378285.1
ENST00000378280.1 ENST00000378288.4 |
TP73
|
tumor protein p73 |
| chr14_+_31343747 | 0.24 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr19_+_13906250 | 0.24 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr12_+_108523133 | 0.24 |
ENST00000547525.1
|
WSCD2
|
WSC domain containing 2 |
| chr5_-_76383133 | 0.23 |
ENST00000255198.2
|
ZBED3
|
zinc finger, BED-type containing 3 |
| chr16_+_67596310 | 0.23 |
ENST00000264010.4
ENST00000401394.1 |
CTCF
|
CCCTC-binding factor (zinc finger protein) |
| chr13_+_25254693 | 0.23 |
ENST00000381946.3
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
| chr13_+_25254545 | 0.22 |
ENST00000218548.6
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
| chr17_+_260097 | 0.22 |
ENST00000360127.6
ENST00000571106.1 ENST00000491373.1 |
C17orf97
|
chromosome 17 open reading frame 97 |
| chr1_+_3569129 | 0.22 |
ENST00000354437.4
ENST00000357733.3 ENST00000346387.4 |
TP73
|
tumor protein p73 |
| chr12_+_108523489 | 0.22 |
ENST00000261400.3
|
WSCD2
|
WSC domain containing 2 |
| chr17_-_15903002 | 0.22 |
ENST00000399277.1
|
ZSWIM7
|
zinc finger, SWIM-type containing 7 |
| chr8_+_67976593 | 0.22 |
ENST00000262210.5
ENST00000412460.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chr16_+_4897912 | 0.21 |
ENST00000545171.1
|
UBN1
|
ubinuclein 1 |
| chr1_+_85527987 | 0.21 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr10_+_45455207 | 0.21 |
ENST00000334940.6
ENST00000374417.2 ENST00000340258.5 ENST00000427758.1 |
RASSF4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr20_-_18038521 | 0.21 |
ENST00000278780.6
|
OVOL2
|
ovo-like zinc finger 2 |
| chr10_-_28287968 | 0.21 |
ENST00000305242.5
|
ARMC4
|
armadillo repeat containing 4 |
| chrX_+_106871713 | 0.21 |
ENST00000372435.4
ENST00000372428.4 ENST00000372419.3 ENST00000543248.1 |
PRPS1
|
phosphoribosyl pyrophosphate synthetase 1 |
| chr1_-_154928562 | 0.20 |
ENST00000368463.3
ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1 |
| chr19_+_33685490 | 0.20 |
ENST00000253193.7
|
LRP3
|
low density lipoprotein receptor-related protein 3 |
| chr10_-_13390270 | 0.20 |
ENST00000378614.4
ENST00000545675.1 ENST00000327347.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chr12_-_48398104 | 0.20 |
ENST00000337299.6
ENST00000380518.3 |
COL2A1
|
collagen, type II, alpha 1 |
| chr4_+_74718906 | 0.20 |
ENST00000226524.3
|
PF4V1
|
platelet factor 4 variant 1 |
| chr11_-_119211525 | 0.20 |
ENST00000528368.1
|
C1QTNF5
|
C1q and tumor necrosis factor related protein 5 |
| chr9_-_130742792 | 0.19 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr9_-_34662651 | 0.19 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr6_+_3849584 | 0.19 |
ENST00000380274.1
ENST00000380272.3 |
FAM50B
|
family with sequence similarity 50, member B |
| chr17_-_43045439 | 0.19 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr10_-_13390021 | 0.19 |
ENST00000537130.1
|
SEPHS1
|
selenophosphate synthetase 1 |
| chr7_+_150758304 | 0.18 |
ENST00000482950.1
ENST00000463414.1 ENST00000310317.5 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr13_+_39261224 | 0.18 |
ENST00000280481.7
|
FREM2
|
FRAS1 related extracellular matrix protein 2 |
| chr17_-_50236039 | 0.18 |
ENST00000451037.2
|
CA10
|
carbonic anhydrase X |
| chr17_-_73178599 | 0.18 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr17_+_38599693 | 0.18 |
ENST00000542955.1
ENST00000269593.4 |
IGFBP4
|
insulin-like growth factor binding protein 4 |
| chr5_+_137688285 | 0.18 |
ENST00000314358.5
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr10_+_134145735 | 0.18 |
ENST00000368613.4
|
LRRC27
|
leucine rich repeat containing 27 |
| chr11_+_73019282 | 0.18 |
ENST00000263674.3
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr20_-_3154162 | 0.18 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr6_+_32006042 | 0.17 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr17_+_38219063 | 0.17 |
ENST00000584985.1
ENST00000264637.4 ENST00000450525.2 |
THRA
|
thyroid hormone receptor, alpha |
| chr6_+_32006159 | 0.17 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr8_-_27462822 | 0.17 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr11_-_118023490 | 0.17 |
ENST00000324727.4
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr16_+_89753070 | 0.17 |
ENST00000353379.7
ENST00000505473.1 ENST00000564192.1 |
CDK10
|
cyclin-dependent kinase 10 |
| chr16_+_30996502 | 0.17 |
ENST00000353250.5
ENST00000262520.6 ENST00000297679.5 ENST00000562932.1 ENST00000574447.1 |
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr10_+_35416223 | 0.16 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr7_-_100808843 | 0.16 |
ENST00000249330.2
|
VGF
|
VGF nerve growth factor inducible |
| chr9_-_98279241 | 0.16 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr6_+_143857949 | 0.16 |
ENST00000367584.4
|
PHACTR2
|
phosphatase and actin regulator 2 |
| chr17_-_50235423 | 0.16 |
ENST00000340813.6
|
CA10
|
carbonic anhydrase X |
| chr8_-_102217796 | 0.16 |
ENST00000519744.1
ENST00000311212.4 ENST00000521272.1 ENST00000519882.1 |
ZNF706
|
zinc finger protein 706 |
| chr1_-_212873267 | 0.16 |
ENST00000243440.1
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr19_-_2721412 | 0.16 |
ENST00000323469.4
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1 |
| chr3_-_112356944 | 0.16 |
ENST00000461431.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr2_+_11273179 | 0.16 |
ENST00000381585.3
ENST00000405022.3 |
C2orf50
|
chromosome 2 open reading frame 50 |
| chr19_-_18385246 | 0.16 |
ENST00000608950.1
ENST00000600328.3 ENST00000600359.3 ENST00000392413.4 |
KIAA1683
|
KIAA1683 |
| chr16_+_4364762 | 0.16 |
ENST00000262366.3
|
GLIS2
|
GLIS family zinc finger 2 |
| chr5_-_179499108 | 0.16 |
ENST00000521389.1
|
RNF130
|
ring finger protein 130 |
| chr19_+_13135731 | 0.15 |
ENST00000587260.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr7_-_100808394 | 0.15 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr19_+_45251804 | 0.15 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr3_-_132441209 | 0.15 |
ENST00000383282.2
ENST00000326682.8 ENST00000343113.4 ENST00000337331.5 |
NPHP3
|
nephronophthisis 3 (adolescent) |
| chr1_+_3569072 | 0.15 |
ENST00000378295.4
ENST00000604074.1 |
TP73
|
tumor protein p73 |
| chr5_-_179499086 | 0.15 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr19_-_12992244 | 0.15 |
ENST00000538460.1
|
DNASE2
|
deoxyribonuclease II, lysosomal |
| chr20_+_30458431 | 0.15 |
ENST00000375938.4
ENST00000535842.1 ENST00000310998.4 ENST00000375921.2 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr11_-_73694346 | 0.15 |
ENST00000310473.3
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr16_-_69364467 | 0.14 |
ENST00000288022.1
|
PDF
|
peptide deformylase (mitochondrial) |
| chr13_+_27131798 | 0.14 |
ENST00000361042.4
|
WASF3
|
WAS protein family, member 3 |
| chr19_+_19174795 | 0.14 |
ENST00000318596.7
|
SLC25A42
|
solute carrier family 25, member 42 |
| chr16_+_29466426 | 0.14 |
ENST00000567248.1
|
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr11_-_407103 | 0.14 |
ENST00000526395.1
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr2_-_98280383 | 0.14 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr17_+_79761997 | 0.14 |
ENST00000400723.3
ENST00000570996.1 |
GCGR
|
glucagon receptor |
| chr3_-_50540854 | 0.14 |
ENST00000423994.2
ENST00000424201.2 ENST00000479441.1 ENST00000429770.1 |
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chrX_-_78622805 | 0.14 |
ENST00000373298.2
|
ITM2A
|
integral membrane protein 2A |
| chr4_+_52917451 | 0.14 |
ENST00000295213.4
ENST00000419395.2 |
SPATA18
|
spermatogenesis associated 18 |
| chr22_-_31328881 | 0.13 |
ENST00000445980.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr19_+_49122548 | 0.13 |
ENST00000245222.4
ENST00000340932.3 ENST00000601712.1 ENST00000600537.1 |
SPHK2
|
sphingosine kinase 2 |
| chr16_+_58059470 | 0.13 |
ENST00000219271.3
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr21_+_46020497 | 0.13 |
ENST00000380102.2
|
KRTAP10-7
|
keratin associated protein 10-7 |
| chr11_-_67275542 | 0.13 |
ENST00000531506.1
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr12_-_59314246 | 0.13 |
ENST00000320743.3
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr7_-_150020750 | 0.13 |
ENST00000539352.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr2_-_56150184 | 0.13 |
ENST00000394554.1
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr16_+_67840986 | 0.13 |
ENST00000561639.1
ENST00000567852.1 ENST00000565148.1 ENST00000388833.3 ENST00000561654.1 ENST00000431934.2 |
TSNAXIP1
|
translin-associated factor X interacting protein 1 |
| chr17_-_80291818 | 0.13 |
ENST00000269389.3
ENST00000581691.1 |
SECTM1
|
secreted and transmembrane 1 |
| chr16_+_67840668 | 0.13 |
ENST00000415766.3
|
TSNAXIP1
|
translin-associated factor X interacting protein 1 |
| chr7_+_150020329 | 0.13 |
ENST00000323078.7
|
LRRC61
|
leucine rich repeat containing 61 |
| chr9_+_214842 | 0.13 |
ENST00000453981.1
ENST00000432829.2 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr10_+_134258649 | 0.12 |
ENST00000392630.3
ENST00000321248.2 |
C10orf91
|
chromosome 10 open reading frame 91 |
| chrX_-_54070607 | 0.12 |
ENST00000338154.6
ENST00000338946.6 |
PHF8
|
PHD finger protein 8 |
| chr12_-_51785182 | 0.12 |
ENST00000356317.3
ENST00000603188.1 ENST00000604847.1 ENST00000604506.1 |
GALNT6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr17_-_73179046 | 0.12 |
ENST00000314523.7
ENST00000420826.2 |
SUMO2
|
small ubiquitin-like modifier 2 |
| chr10_-_100027943 | 0.12 |
ENST00000260702.3
|
LOXL4
|
lysyl oxidase-like 4 |
| chr5_+_121647764 | 0.12 |
ENST00000261368.8
ENST00000379533.2 ENST00000379536.2 ENST00000379538.3 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr1_+_114471809 | 0.12 |
ENST00000426820.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr17_-_4871085 | 0.12 |
ENST00000575142.1
ENST00000206020.3 |
SPAG7
|
sperm associated antigen 7 |
| chr18_+_11752040 | 0.12 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr7_+_150020363 | 0.12 |
ENST00000359623.4
ENST00000493307.1 |
LRRC61
|
leucine rich repeat containing 61 |
| chr19_+_49122785 | 0.12 |
ENST00000598088.1
|
SPHK2
|
sphingosine kinase 2 |
| chr2_+_20646824 | 0.12 |
ENST00000272233.4
|
RHOB
|
ras homolog family member B |
| chr10_+_98592674 | 0.12 |
ENST00000356016.3
ENST00000371097.4 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr2_-_51259229 | 0.12 |
ENST00000405472.3
|
NRXN1
|
neurexin 1 |
| chr6_-_39082922 | 0.12 |
ENST00000229903.4
|
SAYSD1
|
SAYSVFN motif domain containing 1 |
| chr10_+_49514698 | 0.12 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr20_+_60698180 | 0.12 |
ENST00000361670.3
|
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae) |
| chr8_-_41522719 | 0.12 |
ENST00000335651.6
|
ANK1
|
ankyrin 1, erythrocytic |
| chr3_+_184097836 | 0.12 |
ENST00000204604.1
ENST00000310236.3 |
CHRD
|
chordin |
| chr11_+_121322832 | 0.12 |
ENST00000260197.7
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr13_+_27131887 | 0.12 |
ENST00000335327.5
|
WASF3
|
WAS protein family, member 3 |
| chr3_-_134093395 | 0.12 |
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2 |
| chr6_+_134274322 | 0.12 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr4_+_492985 | 0.11 |
ENST00000296306.7
ENST00000536264.1 ENST00000310340.5 ENST00000453061.2 ENST00000504346.1 ENST00000503111.1 ENST00000383028.4 ENST00000509768.1 |
PIGG
|
phosphatidylinositol glycan anchor biosynthesis, class G |
| chr19_-_40724246 | 0.11 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr9_-_74383799 | 0.11 |
ENST00000377044.4
|
TMEM2
|
transmembrane protein 2 |
| chr21_+_43442100 | 0.11 |
ENST00000455701.1
ENST00000596595.1 |
ZNF295-AS1
|
ZNF295 antisense RNA 1 |
| chr15_+_65822756 | 0.11 |
ENST00000562901.1
ENST00000261875.5 ENST00000442729.2 ENST00000565299.1 ENST00000568793.1 |
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr11_-_65150103 | 0.11 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr5_+_121647877 | 0.11 |
ENST00000514497.2
ENST00000261367.7 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr11_-_74109422 | 0.11 |
ENST00000298198.4
|
PGM2L1
|
phosphoglucomutase 2-like 1 |
| chr17_-_3182268 | 0.11 |
ENST00000408891.2
|
OR3A2
|
olfactory receptor, family 3, subfamily A, member 2 |
| chr20_+_49411523 | 0.11 |
ENST00000371608.2
|
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr2_-_202645835 | 0.11 |
ENST00000264276.6
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr17_-_27893990 | 0.11 |
ENST00000307201.4
|
ABHD15
|
abhydrolase domain containing 15 |
| chr17_-_39197699 | 0.11 |
ENST00000306271.4
|
KRTAP1-1
|
keratin associated protein 1-1 |
| chr19_+_10531150 | 0.11 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr15_+_85147127 | 0.11 |
ENST00000541040.1
ENST00000538076.1 ENST00000485222.2 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr1_-_6052463 | 0.11 |
ENST00000378156.4
|
NPHP4
|
nephronophthisis 4 |
| chr19_-_291365 | 0.11 |
ENST00000591572.1
ENST00000269812.3 ENST00000434325.2 |
PPAP2C
|
phosphatidic acid phosphatase type 2C |
| chr21_-_45682099 | 0.11 |
ENST00000270172.3
ENST00000418993.1 |
DNMT3L
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr1_+_38273988 | 0.10 |
ENST00000446260.2
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr16_-_8962544 | 0.10 |
ENST00000570125.1
|
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr3_+_54156570 | 0.10 |
ENST00000415676.2
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr20_-_4982132 | 0.10 |
ENST00000338244.1
ENST00000424750.2 |
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr12_+_3000073 | 0.10 |
ENST00000397132.2
|
TULP3
|
tubby like protein 3 |
| chr21_-_45681765 | 0.10 |
ENST00000431166.1
|
DNMT3L
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr1_+_33352036 | 0.10 |
ENST00000373467.3
|
HPCA
|
hippocalcin |
| chr19_-_1174226 | 0.10 |
ENST00000587024.1
ENST00000361757.3 |
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr19_-_58609570 | 0.10 |
ENST00000600845.1
ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18
|
zinc finger and SCAN domain containing 18 |
| chr16_-_8962200 | 0.10 |
ENST00000562843.1
ENST00000561530.1 ENST00000396593.2 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr17_-_1083078 | 0.10 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chr19_-_12992274 | 0.10 |
ENST00000592506.1
ENST00000222219.3 |
DNASE2
|
deoxyribonuclease II, lysosomal |
| chr16_+_1128781 | 0.10 |
ENST00000293897.4
ENST00000562758.1 |
SSTR5
|
somatostatin receptor 5 |
| chr16_+_4897632 | 0.10 |
ENST00000262376.6
|
UBN1
|
ubinuclein 1 |
| chr15_+_65823092 | 0.10 |
ENST00000566074.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr16_-_8962853 | 0.10 |
ENST00000565287.1
ENST00000311052.5 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC3_ZIC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 0.9 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.2 | 0.5 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.2 | 0.5 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.1 | 0.5 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.4 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.6 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 2.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.3 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.4 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.4 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.4 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.2 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.1 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.1 | 0.4 | GO:0015870 | acetylcholine transport(GO:0015870) |
| 0.1 | 0.2 | GO:0008050 | female courtship behavior(GO:0008050) positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.4 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 1.1 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0080121 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.3 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:1902771 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.4 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.6 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.2 | GO:1902996 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.3 | GO:0060453 | regulation of gastric acid secretion(GO:0060453) |
| 0.0 | 0.0 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.3 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 1.1 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.3 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.0 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.5 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) negative regulation of protein import into nucleus, translocation(GO:0033159) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.0 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.1 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.2 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.0 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.0 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.0 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.5 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 0.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.4 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.3 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.4 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 0.9 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.4 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.5 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 0.4 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.4 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 2.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.2 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.5 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.3 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0008523 | L-ascorbate:sodium symporter activity(GO:0008520) sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 1.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.3 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |