Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
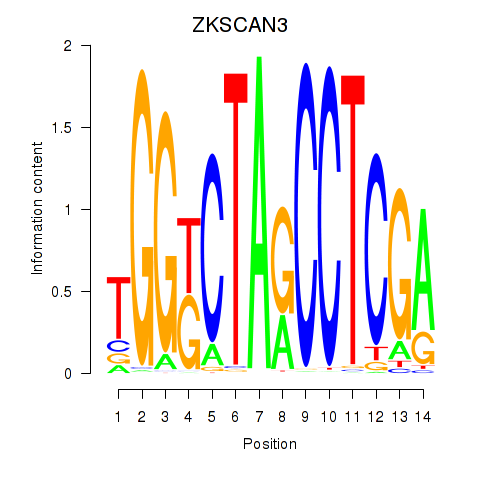
Results for ZKSCAN3
Z-value: 0.36
Transcription factors associated with ZKSCAN3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN3
|
ENSG00000189298.9 | zinc finger with KRAB and SCAN domains 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZKSCAN3 | hg19_v2_chr6_+_28317685_28317761 | 0.19 | 3.2e-01 | Click! |
Activity profile of ZKSCAN3 motif
Sorted Z-values of ZKSCAN3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_3598871 | 1.68 |
ENST00000603362.1
ENST00000604479.1 |
TP73
|
tumor protein p73 |
| chr9_+_124926856 | 1.14 |
ENST00000418632.1
|
MORN5
|
MORN repeat containing 5 |
| chr15_-_41624685 | 1.14 |
ENST00000560640.1
ENST00000220514.3 |
OIP5
|
Opa interacting protein 5 |
| chr15_+_41624892 | 0.93 |
ENST00000260359.6
ENST00000450318.1 ENST00000450592.2 ENST00000559596.1 ENST00000414849.2 ENST00000560747.1 ENST00000560177.1 |
NUSAP1
|
nucleolar and spindle associated protein 1 |
| chr19_+_39786962 | 0.64 |
ENST00000333625.2
|
IFNL1
|
interferon, lambda 1 |
| chr18_-_74728998 | 0.43 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr19_+_39759154 | 0.43 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr19_-_39735646 | 0.40 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr16_+_2479390 | 0.30 |
ENST00000397066.4
|
CCNF
|
cyclin F |
| chr14_-_106926724 | 0.28 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr7_+_155089486 | 0.28 |
ENST00000340368.4
ENST00000344756.4 ENST00000425172.1 ENST00000342407.5 |
INSIG1
|
insulin induced gene 1 |
| chr2_+_191745560 | 0.28 |
ENST00000338435.4
|
GLS
|
glutaminase |
| chr9_+_138235095 | 0.27 |
ENST00000320778.2
|
C9orf62
|
chromosome 9 open reading frame 62 |
| chr5_+_65018017 | 0.24 |
ENST00000380985.5
ENST00000502464.1 |
NLN
|
neurolysin (metallopeptidase M3 family) |
| chr10_+_6779326 | 0.23 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr15_+_74610894 | 0.20 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr14_-_80697396 | 0.20 |
ENST00000557010.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr10_+_21823079 | 0.19 |
ENST00000377100.3
ENST00000377072.3 ENST00000446906.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr7_+_155090271 | 0.19 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr14_-_106069247 | 0.17 |
ENST00000479229.1
|
RP11-731F5.1
|
RP11-731F5.1 |
| chr11_-_64013288 | 0.15 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr7_-_155089251 | 0.15 |
ENST00000609974.1
|
AC144652.1
|
AC144652.1 |
| chr19_-_42348692 | 0.14 |
ENST00000330743.3
ENST00000601246.1 |
LYPD4
|
LY6/PLAUR domain containing 4 |
| chr10_+_21823243 | 0.14 |
ENST00000307729.7
ENST00000377091.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr22_+_50312316 | 0.14 |
ENST00000328268.4
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr22_-_50312052 | 0.13 |
ENST00000330817.6
|
ALG12
|
ALG12, alpha-1,6-mannosyltransferase |
| chrX_+_153775821 | 0.12 |
ENST00000263518.6
ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr8_+_123793633 | 0.11 |
ENST00000314393.4
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr11_-_64013663 | 0.11 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr22_+_50312379 | 0.10 |
ENST00000407217.3
ENST00000403427.3 |
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr6_-_31508304 | 0.10 |
ENST00000376177.2
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr12_-_3982511 | 0.10 |
ENST00000427057.2
ENST00000228820.4 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr5_-_65017921 | 0.10 |
ENST00000381007.4
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr22_+_50312274 | 0.10 |
ENST00000404488.3
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr12_-_3982548 | 0.09 |
ENST00000397096.2
ENST00000447133.3 ENST00000450737.2 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr1_-_115259337 | 0.08 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr1_+_93913713 | 0.08 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chrX_-_135338503 | 0.08 |
ENST00000370663.5
|
MAP7D3
|
MAP7 domain containing 3 |
| chr16_-_745946 | 0.07 |
ENST00000562563.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chrX_-_153775426 | 0.07 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr19_-_49258606 | 0.06 |
ENST00000310160.3
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) |
| chr1_+_93913665 | 0.06 |
ENST00000271234.7
ENST00000370256.4 ENST00000260506.8 |
FNBP1L
|
formin binding protein 1-like |
| chr3_-_122512619 | 0.06 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr1_-_28415075 | 0.06 |
ENST00000373863.3
ENST00000436342.2 ENST00000540618.1 ENST00000545175.1 |
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr12_+_109535373 | 0.06 |
ENST00000242576.2
|
UNG
|
uracil-DNA glycosylase |
| chr1_-_19811996 | 0.06 |
ENST00000264203.3
ENST00000401084.2 |
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr19_+_32836499 | 0.04 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr1_+_226250379 | 0.04 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr1_-_19811962 | 0.03 |
ENST00000375142.1
ENST00000264202.6 |
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr5_+_140079919 | 0.03 |
ENST00000274712.3
|
ZMAT2
|
zinc finger, matrin-type 2 |
| chr14_+_94492674 | 0.03 |
ENST00000203664.5
ENST00000553723.1 |
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr12_+_100867694 | 0.03 |
ENST00000392986.3
ENST00000549996.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr5_-_95297534 | 0.02 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr1_-_28415204 | 0.02 |
ENST00000373871.3
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr5_-_114598548 | 0.01 |
ENST00000379615.3
ENST00000419445.1 |
PGGT1B
|
protein geranylgeranyltransferase type I, beta subunit |
| chr1_-_99470368 | 0.01 |
ENST00000263177.4
|
LPPR5
|
Lipid phosphate phosphatase-related protein type 5 |
| chr17_-_60142609 | 0.00 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr12_+_109535923 | 0.00 |
ENST00000336865.2
|
UNG
|
uracil-DNA glycosylase |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZKSCAN3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 1.7 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.1 | 0.6 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 0.4 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.2 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 1.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.3 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.4 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.0 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 1.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |