Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
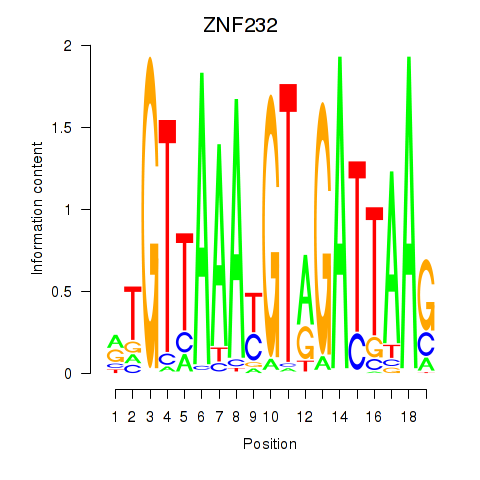
Results for ZNF232
Z-value: 0.41
Transcription factors associated with ZNF232
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF232
|
ENSG00000167840.9 | zinc finger protein 232 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF232 | hg19_v2_chr17_-_5015129_5015187 | 0.43 | 1.7e-02 | Click! |
Activity profile of ZNF232 motif
Sorted Z-values of ZNF232 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_228735763 | 2.26 |
ENST00000373666.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chrY_+_15016725 | 1.33 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr15_-_56757329 | 1.22 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr10_+_134150835 | 1.15 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr7_+_142829162 | 0.90 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr7_-_56101826 | 0.88 |
ENST00000421626.1
|
PSPH
|
phosphoserine phosphatase |
| chrY_+_15016013 | 0.76 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr3_+_160559931 | 0.59 |
ENST00000464260.1
ENST00000295839.9 |
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr17_+_26800648 | 0.58 |
ENST00000545060.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr19_-_11545920 | 0.58 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr17_+_26800756 | 0.57 |
ENST00000537681.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr17_+_26800296 | 0.51 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr3_+_8543533 | 0.45 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr1_+_182419261 | 0.37 |
ENST00000294854.8
ENST00000542961.1 |
RGSL1
|
regulator of G-protein signaling like 1 |
| chr1_+_174669653 | 0.35 |
ENST00000325589.5
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr7_+_116654935 | 0.30 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr4_+_185570871 | 0.27 |
ENST00000512834.1
|
PRIMPOL
|
primase and polymerase (DNA-directed) |
| chr10_-_61900762 | 0.26 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chrX_+_36065053 | 0.23 |
ENST00000313548.4
|
CHDC2
|
calponin homology domain containing 2 |
| chr6_-_26056695 | 0.22 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr3_+_8543393 | 0.20 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr3_+_8543561 | 0.19 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr15_-_60771128 | 0.17 |
ENST00000558512.1
ENST00000561114.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr2_-_113522177 | 0.16 |
ENST00000541405.1
|
CKAP2L
|
cytoskeleton associated protein 2-like |
| chr21_+_38792602 | 0.16 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr15_-_60771280 | 0.16 |
ENST00000560072.1
ENST00000560406.1 ENST00000560520.1 ENST00000261520.4 ENST00000439632.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr6_+_109416684 | 0.15 |
ENST00000521522.1
ENST00000524064.1 ENST00000522608.1 ENST00000521503.1 ENST00000519407.1 ENST00000519095.1 ENST00000368968.2 ENST00000522490.1 ENST00000523209.1 ENST00000368970.2 ENST00000520883.1 ENST00000523787.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr11_+_66276550 | 0.13 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr10_-_73533255 | 0.13 |
ENST00000394957.3
|
C10orf54
|
chromosome 10 open reading frame 54 |
| chr4_-_88450612 | 0.12 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr14_-_21516590 | 0.12 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chr7_+_1609694 | 0.11 |
ENST00000437621.2
ENST00000457484.2 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr1_+_84944926 | 0.10 |
ENST00000370656.1
ENST00000370654.5 |
RPF1
|
ribosome production factor 1 homolog (S. cerevisiae) |
| chr6_+_109416313 | 0.09 |
ENST00000521277.1
ENST00000517392.1 ENST00000407272.1 ENST00000336977.4 ENST00000519286.1 ENST00000518329.1 ENST00000522461.1 ENST00000518853.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr1_-_16556038 | 0.09 |
ENST00000375605.2
|
C1orf134
|
chromosome 1 open reading frame 134 |
| chr1_+_237205476 | 0.09 |
ENST00000366574.2
|
RYR2
|
ryanodine receptor 2 (cardiac) |
| chr12_-_9885888 | 0.08 |
ENST00000327839.3
|
CLECL1
|
C-type lectin-like 1 |
| chr1_+_40997233 | 0.08 |
ENST00000372699.3
ENST00000372697.3 ENST00000372696.3 |
ZNF684
|
zinc finger protein 684 |
| chr1_-_204654826 | 0.08 |
ENST00000367177.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr2_-_113522248 | 0.08 |
ENST00000302450.6
|
CKAP2L
|
cytoskeleton associated protein 2-like |
| chr2_+_74425689 | 0.08 |
ENST00000394053.2
ENST00000409804.1 ENST00000264090.4 ENST00000394050.3 ENST00000409601.1 |
MTHFD2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr18_+_61637159 | 0.07 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr4_-_84255935 | 0.07 |
ENST00000513463.1
|
HPSE
|
heparanase |
| chr16_-_20367584 | 0.06 |
ENST00000570689.1
|
UMOD
|
uromodulin |
| chr3_+_51863433 | 0.05 |
ENST00000444293.1
|
IQCF3
|
IQ motif containing F3 |
| chr1_-_160549235 | 0.05 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr11_-_18127566 | 0.05 |
ENST00000532452.1
ENST00000530180.1 ENST00000300013.4 ENST00000529318.1 ENST00000524803.1 |
SAAL1
|
serum amyloid A-like 1 |
| chr6_+_24403144 | 0.04 |
ENST00000274747.7
ENST00000543597.1 ENST00000535061.1 ENST00000378353.1 ENST00000378386.3 ENST00000443868.2 |
MRS2
|
MRS2 magnesium transporter |
| chr7_+_142919130 | 0.04 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr5_-_180076613 | 0.03 |
ENST00000261937.6
ENST00000393347.3 |
FLT4
|
fms-related tyrosine kinase 4 |
| chr2_+_241807870 | 0.03 |
ENST00000307503.3
|
AGXT
|
alanine-glyoxylate aminotransferase |
| chr10_-_27389392 | 0.03 |
ENST00000376087.4
|
ANKRD26
|
ankyrin repeat domain 26 |
| chr1_-_53686261 | 0.02 |
ENST00000294360.4
|
C1orf123
|
chromosome 1 open reading frame 123 |
| chr19_-_38210622 | 0.02 |
ENST00000591664.1
ENST00000355202.4 |
ZNF607
|
zinc finger protein 607 |
| chr17_-_6915646 | 0.01 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chrX_-_100548045 | 0.01 |
ENST00000372907.3
ENST00000372905.2 |
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF232
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.3 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 2.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0098907 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.9 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 2.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 1.7 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.1 | 0.9 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 1.0 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 2.1 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |