Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
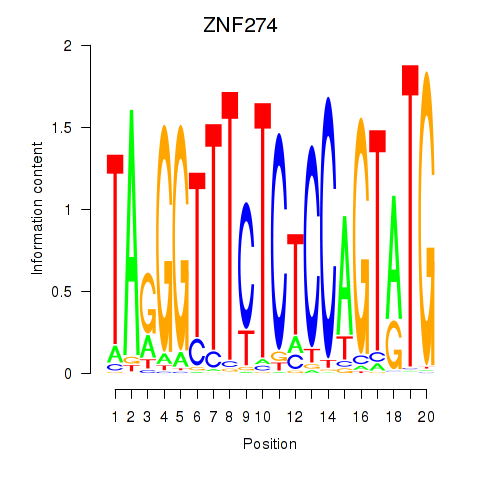
Results for ZNF274
Z-value: 0.33
Transcription factors associated with ZNF274
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF274
|
ENSG00000171606.13 | zinc finger protein 274 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF274 | hg19_v2_chr19_+_58694396_58694485 | -0.22 | 2.5e-01 | Click! |
Activity profile of ZNF274 motif
Sorted Z-values of ZNF274 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_45205498 | 0.77 |
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C |
| chr1_+_45205478 | 0.77 |
ENST00000452259.1
ENST00000372224.4 |
KIF2C
|
kinesin family member 2C |
| chr2_+_11864458 | 0.38 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr6_+_31553901 | 0.37 |
ENST00000418507.2
ENST00000438075.2 ENST00000376100.3 ENST00000376111.4 |
LST1
|
leukocyte specific transcript 1 |
| chr12_-_54653313 | 0.37 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr6_+_31553978 | 0.34 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr16_+_29818857 | 0.33 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr6_+_30687978 | 0.33 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr2_-_42991257 | 0.32 |
ENST00000378661.2
|
OXER1
|
oxoeicosanoid (OXE) receptor 1 |
| chr22_+_39353527 | 0.28 |
ENST00000249116.2
|
APOBEC3A
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
| chr17_-_41132410 | 0.27 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr14_+_22409308 | 0.24 |
ENST00000390441.2
|
TRAV9-2
|
T cell receptor alpha variable 9-2 |
| chr6_-_32784687 | 0.24 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr18_+_9885760 | 0.24 |
ENST00000536353.2
ENST00000584255.1 |
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr12_+_109273806 | 0.23 |
ENST00000228476.3
ENST00000547768.1 |
DAO
|
D-amino-acid oxidase |
| chr3_+_102153859 | 0.21 |
ENST00000306176.1
ENST00000466937.1 |
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr19_+_40195101 | 0.20 |
ENST00000360675.3
ENST00000601802.1 |
LGALS14
|
lectin, galactoside-binding, soluble, 14 |
| chr20_+_60174827 | 0.20 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr12_+_113344755 | 0.20 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr11_-_10590118 | 0.19 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr6_+_36165133 | 0.19 |
ENST00000446974.1
ENST00000454960.1 |
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr11_+_55653396 | 0.18 |
ENST00000244891.3
|
TRIM51
|
tripartite motif-containing 51 |
| chr8_-_4852494 | 0.18 |
ENST00000520002.1
ENST00000602557.1 |
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr8_-_4852218 | 0.16 |
ENST00000400186.3
ENST00000602723.1 |
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr10_-_29811456 | 0.16 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr16_-_80603558 | 0.16 |
ENST00000567317.1
|
RP11-18F14.1
|
RP11-18F14.1 |
| chr19_+_6135646 | 0.15 |
ENST00000588304.1
ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr16_+_29819096 | 0.15 |
ENST00000568411.1
ENST00000563012.1 ENST00000562557.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_-_89292422 | 0.15 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr18_+_9885961 | 0.14 |
ENST00000306084.6
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr19_+_40194946 | 0.14 |
ENST00000392052.3
|
LGALS14
|
lectin, galactoside-binding, soluble, 14 |
| chr15_+_89164520 | 0.14 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chr19_+_6887571 | 0.13 |
ENST00000250572.8
ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr14_-_24047965 | 0.11 |
ENST00000397118.3
ENST00000356300.4 |
JPH4
|
junctophilin 4 |
| chr5_+_32531893 | 0.11 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr10_+_5488564 | 0.11 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr16_+_29819372 | 0.11 |
ENST00000568544.1
ENST00000569978.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr5_-_74162605 | 0.10 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr18_+_9885934 | 0.10 |
ENST00000357775.5
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr16_+_29819446 | 0.10 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chrX_+_52920336 | 0.08 |
ENST00000452154.2
|
FAM156B
|
family with sequence similarity 156, member B |
| chr7_+_30791743 | 0.06 |
ENST00000013222.5
ENST00000409539.1 |
INMT
|
indolethylamine N-methyltransferase |
| chrX_+_119737806 | 0.05 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr3_+_50284321 | 0.05 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr19_+_41949054 | 0.05 |
ENST00000378187.2
|
C19orf69
|
chromosome 19 open reading frame 69 |
| chr16_+_31470179 | 0.03 |
ENST00000538189.1
ENST00000268314.4 |
ARMC5
|
armadillo repeat containing 5 |
| chr17_-_3461092 | 0.02 |
ENST00000301365.4
ENST00000572519.1 |
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr2_+_9696028 | 0.02 |
ENST00000607241.1
|
RP11-214N9.1
|
RP11-214N9.1 |
| chr2_+_178977143 | 0.01 |
ENST00000286070.5
|
RBM45
|
RNA binding motif protein 45 |
| chr19_-_19774473 | 0.01 |
ENST00000357324.6
|
ATP13A1
|
ATPase type 13A1 |
| chr1_+_42928945 | 0.01 |
ENST00000428554.2
|
CCDC30
|
coiled-coil domain containing 30 |
| chr4_-_121993673 | 0.00 |
ENST00000379692.4
|
NDNF
|
neuron-derived neurotrophic factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF274
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.2 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.3 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.5 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.3 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.3 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |