Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
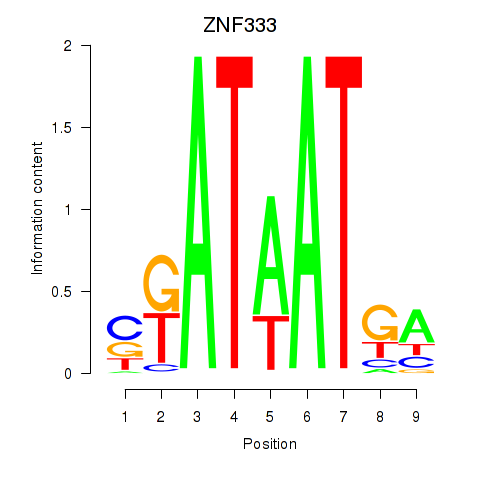
Results for ZNF333
Z-value: 0.64
Transcription factors associated with ZNF333
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF333
|
ENSG00000160961.7 | zinc finger protein 333 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF333 | hg19_v2_chr19_+_14800711_14800917 | 0.33 | 7.9e-02 | Click! |
Activity profile of ZNF333 motif
Sorted Z-values of ZNF333 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_112693865 | 2.44 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr7_-_6006768 | 1.76 |
ENST00000441023.2
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr7_+_6796986 | 1.66 |
ENST00000297186.3
|
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr7_-_6007070 | 1.64 |
ENST00000337579.3
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr3_-_112564797 | 1.60 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr9_-_39239171 | 1.32 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr7_+_6797288 | 1.23 |
ENST00000433859.2
ENST00000359718.3 |
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr5_-_138718973 | 1.11 |
ENST00000353963.3
ENST00000348729.3 |
SLC23A1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr6_+_29274403 | 0.90 |
ENST00000377160.2
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1 |
| chr2_+_233527443 | 0.87 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr12_-_71031185 | 0.67 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr3_+_159557637 | 0.66 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr1_-_197036364 | 0.66 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr3_+_35721182 | 0.62 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr4_+_66536248 | 0.61 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr7_+_141463897 | 0.59 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr9_+_133971909 | 0.58 |
ENST00000247291.3
ENST00000372302.1 ENST00000372300.1 ENST00000372298.1 |
AIF1L
|
allograft inflammatory factor 1-like |
| chr9_+_133971863 | 0.58 |
ENST00000372309.3
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr10_+_45495898 | 0.54 |
ENST00000298299.3
|
ZNF22
|
zinc finger protein 22 |
| chr12_-_100656134 | 0.52 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr7_-_105319536 | 0.51 |
ENST00000477775.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr3_+_35721106 | 0.46 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr12_-_87232644 | 0.44 |
ENST00000549405.2
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr7_-_130080977 | 0.42 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr17_+_4613776 | 0.40 |
ENST00000269260.2
|
ARRB2
|
arrestin, beta 2 |
| chr13_-_114107839 | 0.39 |
ENST00000375418.3
|
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr14_+_57671888 | 0.39 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr17_-_33446735 | 0.37 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr5_-_16509101 | 0.37 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr10_+_118350468 | 0.36 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chrX_-_114253536 | 0.34 |
ENST00000371936.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr3_-_126327398 | 0.34 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr12_-_71031220 | 0.33 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr7_-_25268104 | 0.32 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr7_-_83824169 | 0.30 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_+_4613918 | 0.29 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr10_+_118350522 | 0.28 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr19_-_10446449 | 0.28 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr4_-_100356551 | 0.28 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr17_-_60883993 | 0.27 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr4_+_71108300 | 0.27 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr11_+_92085262 | 0.27 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr1_-_153123345 | 0.27 |
ENST00000368748.4
|
SPRR2G
|
small proline-rich protein 2G |
| chrY_-_6742068 | 0.26 |
ENST00000215479.5
|
AMELY
|
amelogenin, Y-linked |
| chr1_-_36789755 | 0.26 |
ENST00000270824.1
|
EVA1B
|
eva-1 homolog B (C. elegans) |
| chr5_+_137203465 | 0.26 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr16_+_84328252 | 0.25 |
ENST00000219454.5
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr4_+_110749143 | 0.25 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr16_+_84328429 | 0.23 |
ENST00000568638.1
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr4_-_26492076 | 0.23 |
ENST00000295589.3
|
CCKAR
|
cholecystokinin A receptor |
| chr19_-_45004556 | 0.23 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr3_+_173116225 | 0.23 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr2_+_191002486 | 0.22 |
ENST00000396974.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr8_-_114449112 | 0.22 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chrX_-_107975917 | 0.22 |
ENST00000563887.1
|
RP6-24A23.6
|
Uncharacterized protein |
| chr11_-_72504637 | 0.20 |
ENST00000536377.1
ENST00000359373.5 |
STARD10
ARAP1
|
StAR-related lipid transfer (START) domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr10_-_23633720 | 0.20 |
ENST00000323327.4
|
C10orf67
|
chromosome 10 open reading frame 67 |
| chr16_+_87636474 | 0.20 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr15_+_54305101 | 0.20 |
ENST00000260323.11
ENST00000545554.1 ENST00000537900.1 |
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr6_+_160327974 | 0.18 |
ENST00000252660.4
|
MAS1
|
MAS1 oncogene |
| chr18_+_28956740 | 0.18 |
ENST00000308128.4
ENST00000359747.4 |
DSG4
|
desmoglein 4 |
| chr7_-_36406750 | 0.18 |
ENST00000453212.1
ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895
|
KIAA0895 |
| chr15_+_36338242 | 0.17 |
ENST00000560056.1
|
RP11-684B21.1
|
RP11-684B21.1 |
| chr12_+_58176525 | 0.17 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr11_-_6440624 | 0.16 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr9_+_109625378 | 0.16 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chrX_+_99839799 | 0.16 |
ENST00000373031.4
|
TNMD
|
tenomodulin |
| chr7_-_115670804 | 0.15 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr9_-_19786926 | 0.15 |
ENST00000341998.2
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr7_-_115670792 | 0.14 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr20_-_44600810 | 0.14 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chrX_+_11311533 | 0.14 |
ENST00000380714.3
ENST00000380712.3 ENST00000348912.4 |
AMELX
|
amelogenin, X-linked |
| chr12_+_109785708 | 0.14 |
ENST00000310903.5
|
MYO1H
|
myosin IH |
| chr2_-_128432639 | 0.14 |
ENST00000545738.2
ENST00000355119.4 ENST00000409808.2 |
LIMS2
|
LIM and senescent cell antigen-like domains 2 |
| chr1_-_169599314 | 0.13 |
ENST00000367786.2
ENST00000458599.2 ENST00000367795.2 ENST00000263686.6 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr1_-_169599353 | 0.13 |
ENST00000367793.2
ENST00000367794.2 ENST00000367792.2 ENST00000367791.2 ENST00000367788.2 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr3_+_174577070 | 0.12 |
ENST00000454872.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr3_-_125094093 | 0.12 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr11_+_12766583 | 0.12 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr10_-_32667660 | 0.12 |
ENST00000375110.2
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr15_+_69857515 | 0.11 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr19_-_42721819 | 0.11 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr3_+_38323785 | 0.11 |
ENST00000466887.1
ENST00000448498.1 |
SLC22A14
|
solute carrier family 22, member 14 |
| chr18_-_40857493 | 0.10 |
ENST00000255224.3
|
SYT4
|
synaptotagmin IV |
| chr10_-_128210005 | 0.10 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr17_-_3337135 | 0.10 |
ENST00000248384.1
|
OR1E2
|
olfactory receptor, family 1, subfamily E, member 2 |
| chr6_+_50786414 | 0.10 |
ENST00000344788.3
ENST00000393655.3 ENST00000263046.4 |
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr14_+_85996507 | 0.10 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr20_+_58515417 | 0.10 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr17_-_33390667 | 0.10 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr3_+_101659682 | 0.10 |
ENST00000465215.1
|
RP11-221J22.1
|
RP11-221J22.1 |
| chr11_-_71823796 | 0.10 |
ENST00000545680.1
ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr12_-_122879969 | 0.10 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr6_-_32908792 | 0.09 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr11_-_71823715 | 0.09 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr20_+_56964169 | 0.09 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr10_+_121410882 | 0.09 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chrX_-_119445306 | 0.08 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr15_+_38746307 | 0.08 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
| chr6_+_131894284 | 0.08 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr14_+_85996471 | 0.07 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr10_+_90346519 | 0.07 |
ENST00000371939.3
|
LIPJ
|
lipase, family member J |
| chr17_-_57229155 | 0.06 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr14_+_76618242 | 0.06 |
ENST00000557542.1
ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L
|
G patch domain containing 2-like |
| chr5_-_78281775 | 0.06 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chrX_-_21776281 | 0.06 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr5_-_35195338 | 0.06 |
ENST00000509839.1
|
PRLR
|
prolactin receptor |
| chr8_-_139926236 | 0.05 |
ENST00000303045.6
ENST00000435777.1 |
COL22A1
|
collagen, type XXII, alpha 1 |
| chr6_-_2634820 | 0.05 |
ENST00000296847.3
|
C6orf195
|
chromosome 6 open reading frame 195 |
| chr4_-_186696425 | 0.05 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_+_107318157 | 0.04 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr20_+_1099233 | 0.04 |
ENST00000246015.4
ENST00000335877.6 ENST00000438768.2 |
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr11_+_74303575 | 0.04 |
ENST00000263681.2
|
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr11_-_18062872 | 0.03 |
ENST00000250018.2
|
TPH1
|
tryptophan hydroxylase 1 |
| chr14_+_105046094 | 0.03 |
ENST00000331952.2
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr17_+_33288549 | 0.02 |
ENST00000361952.3
|
ZNF830
|
zinc finger protein 830 |
| chr11_-_72504681 | 0.02 |
ENST00000538536.1
ENST00000543304.1 ENST00000540587.1 ENST00000334805.6 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr5_+_173315283 | 0.02 |
ENST00000265085.5
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr5_+_137225125 | 0.02 |
ENST00000350250.4
ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr12_-_7656357 | 0.02 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chr20_+_56964253 | 0.01 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr2_-_178753465 | 0.01 |
ENST00000389683.3
|
PDE11A
|
phosphodiesterase 11A |
| chr2_+_217363559 | 0.01 |
ENST00000600880.1
ENST00000446558.1 |
RPL37A
|
ribosomal protein L37a |
| chr12_-_10251539 | 0.01 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr14_+_105046021 | 0.01 |
ENST00000557649.1
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr1_+_95975672 | 0.00 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr2_+_185463093 | 0.00 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr2_-_145275228 | 0.00 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr20_-_33413416 | 0.00 |
ENST00000359003.2
|
NCOA6
|
nuclear receptor coactivator 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF333
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.7 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.2 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 0.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.3 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 0.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.3 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.3 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.3 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.3 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.2 | 0.7 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 0.4 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.2 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 0.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.4 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 2.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |