Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
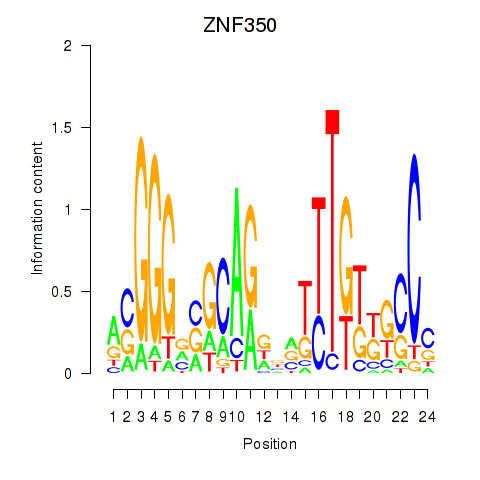
Results for ZNF350
Z-value: 0.55
Transcription factors associated with ZNF350
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF350
|
ENSG00000256683.2 | zinc finger protein 350 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF350 | hg19_v2_chr19_-_52489923_52490113 | 0.41 | 2.6e-02 | Click! |
Activity profile of ZNF350 motif
Sorted Z-values of ZNF350 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_132286754 | 3.45 |
ENST00000434330.1
|
CCDC74A
|
coiled-coil domain containing 74A |
| chr2_+_132287237 | 2.89 |
ENST00000467992.2
|
CCDC74A
|
coiled-coil domain containing 74A |
| chr1_-_46089718 | 2.32 |
ENST00000421127.2
ENST00000343901.2 ENST00000528266.1 |
CCDC17
|
coiled-coil domain containing 17 |
| chr17_+_68071389 | 2.17 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_+_100328433 | 2.17 |
ENST00000273352.3
|
GPR128
|
G protein-coupled receptor 128 |
| chr17_+_68071458 | 2.15 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr4_+_75858290 | 2.02 |
ENST00000513238.1
|
PARM1
|
prostate androgen-regulated mucin-like protein 1 |
| chr4_+_75858318 | 1.83 |
ENST00000307428.7
|
PARM1
|
prostate androgen-regulated mucin-like protein 1 |
| chr1_-_46089639 | 1.75 |
ENST00000445048.2
|
CCDC17
|
coiled-coil domain containing 17 |
| chr1_-_98510843 | 1.47 |
ENST00000413670.2
ENST00000538428.1 |
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr16_+_69600209 | 1.36 |
ENST00000566899.1
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr17_+_68165657 | 1.26 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr20_+_62367989 | 1.25 |
ENST00000309546.3
|
LIME1
|
Lck interacting transmembrane adaptor 1 |
| chr5_+_42423872 | 1.24 |
ENST00000230882.4
ENST00000357703.3 |
GHR
|
growth hormone receptor |
| chr2_+_233497931 | 1.20 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr11_-_63381925 | 1.20 |
ENST00000415826.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr16_+_69600058 | 1.15 |
ENST00000393742.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr11_-_45687128 | 0.81 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr2_-_157198860 | 0.78 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr2_+_120301997 | 0.78 |
ENST00000602047.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr2_+_120302041 | 0.77 |
ENST00000442513.3
ENST00000413369.3 |
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr14_+_96671016 | 0.74 |
ENST00000542454.2
ENST00000554311.1 ENST00000306005.3 ENST00000539359.1 ENST00000553811.1 |
BDKRB2
RP11-404P21.8
|
bradykinin receptor B2 Uncharacterized protein |
| chr4_-_76008706 | 0.73 |
ENST00000562355.1
ENST00000563602.1 |
RP11-44F21.5
|
RP11-44F21.5 |
| chr18_+_77155856 | 0.71 |
ENST00000253506.5
ENST00000591814.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr12_+_50451331 | 0.71 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr12_+_50451462 | 0.69 |
ENST00000447966.2
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr11_-_62473776 | 0.64 |
ENST00000278893.7
ENST00000407022.3 ENST00000421906.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr3_-_113775328 | 0.63 |
ENST00000483766.1
ENST00000545063.1 ENST00000491000.1 ENST00000295878.3 |
KIAA1407
|
KIAA1407 |
| chr13_-_96329048 | 0.63 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
| chr17_+_7487146 | 0.58 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr11_-_62473706 | 0.57 |
ENST00000403550.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr11_-_8954491 | 0.56 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr12_+_81471816 | 0.54 |
ENST00000261206.3
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chr1_-_33647267 | 0.49 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr8_+_86157699 | 0.49 |
ENST00000321764.3
|
CA13
|
carbonic anhydrase XIII |
| chr11_-_414948 | 0.48 |
ENST00000530494.1
ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr11_-_63381823 | 0.45 |
ENST00000323646.5
|
PLA2G16
|
phospholipase A2, group XVI |
| chr11_-_327537 | 0.43 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr4_+_6784401 | 0.41 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr3_-_11623804 | 0.40 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr6_-_56707943 | 0.40 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr10_-_12084770 | 0.39 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr16_+_71879861 | 0.37 |
ENST00000427980.2
ENST00000568581.1 |
ATXN1L
IST1
|
ataxin 1-like increased sodium tolerance 1 homolog (yeast) |
| chr3_+_140947563 | 0.35 |
ENST00000505013.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr18_+_77155942 | 0.34 |
ENST00000397790.2
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr12_+_131438443 | 0.34 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr10_+_69644404 | 0.34 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr11_-_119217365 | 0.32 |
ENST00000360167.4
ENST00000555262.1 ENST00000449574.2 ENST00000445041.2 |
MFRP
C1QTNF5
|
membrane frizzled-related protein C1q and tumor necrosis factor related protein 5 |
| chr16_+_14802801 | 0.32 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr17_+_6554971 | 0.32 |
ENST00000391428.2
|
C17orf100
|
chromosome 17 open reading frame 100 |
| chr7_+_2559399 | 0.29 |
ENST00000222725.5
ENST00000359574.3 |
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr11_+_64323428 | 0.27 |
ENST00000377581.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr8_+_144295067 | 0.26 |
ENST00000330824.2
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr11_-_7041466 | 0.26 |
ENST00000536068.1
ENST00000278314.4 |
ZNF214
|
zinc finger protein 214 |
| chr11_-_9113093 | 0.24 |
ENST00000450649.2
|
SCUBE2
|
signal peptide, CUB domain, EGF-like 2 |
| chr5_-_107717058 | 0.21 |
ENST00000359660.5
|
FBXL17
|
F-box and leucine-rich repeat protein 17 |
| chr10_-_94003003 | 0.21 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr16_+_31225337 | 0.21 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr11_-_9113137 | 0.20 |
ENST00000520467.1
ENST00000309263.3 ENST00000457346.2 |
SCUBE2
|
signal peptide, CUB domain, EGF-like 2 |
| chr15_+_57210818 | 0.20 |
ENST00000438423.2
ENST00000267811.5 ENST00000452095.2 ENST00000559609.1 ENST00000333725.5 |
TCF12
|
transcription factor 12 |
| chr11_-_64612041 | 0.19 |
ENST00000342711.5
|
CDC42BPG
|
CDC42 binding protein kinase gamma (DMPK-like) |
| chr1_-_51425902 | 0.19 |
ENST00000396153.2
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr17_+_19030782 | 0.18 |
ENST00000344415.4
ENST00000577213.1 |
GRAPL
|
GRB2-related adaptor protein-like |
| chr6_-_28303901 | 0.18 |
ENST00000439158.1
ENST00000446474.1 ENST00000414431.1 ENST00000344279.6 ENST00000453745.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr12_-_53228079 | 0.17 |
ENST00000330553.5
|
KRT79
|
keratin 79 |
| chr14_+_73603126 | 0.17 |
ENST00000557356.1
ENST00000556864.1 ENST00000556533.1 ENST00000556951.1 ENST00000557293.1 ENST00000553719.1 ENST00000553599.1 ENST00000556011.1 ENST00000394157.3 ENST00000357710.4 ENST00000324501.5 ENST00000560005.2 ENST00000555254.1 ENST00000261970.3 ENST00000344094.3 ENST00000554131.1 ENST00000557037.1 |
PSEN1
|
presenilin 1 |
| chr19_+_42829702 | 0.17 |
ENST00000334370.4
|
MEGF8
|
multiple EGF-like-domains 8 |
| chr4_+_108910870 | 0.17 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr8_-_12612962 | 0.17 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr11_-_64511789 | 0.16 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr4_+_108911036 | 0.15 |
ENST00000505878.1
|
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr16_+_67282853 | 0.13 |
ENST00000299798.11
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr1_+_228395755 | 0.13 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr1_+_204797749 | 0.12 |
ENST00000367172.4
ENST00000367171.4 ENST00000367170.4 ENST00000338515.6 ENST00000339876.6 ENST00000338586.6 ENST00000539706.1 ENST00000360049.4 ENST00000367169.4 ENST00000446412.1 ENST00000403080.1 |
NFASC
|
neurofascin |
| chr15_+_57210961 | 0.12 |
ENST00000557843.1
|
TCF12
|
transcription factor 12 |
| chr1_-_247094628 | 0.12 |
ENST00000366508.1
ENST00000326225.3 ENST00000391829.2 |
AHCTF1
|
AT hook containing transcription factor 1 |
| chr12_+_54519842 | 0.11 |
ENST00000508564.1
|
RP11-834C11.4
|
RP11-834C11.4 |
| chrX_-_17878827 | 0.11 |
ENST00000360011.1
|
RAI2
|
retinoic acid induced 2 |
| chr1_+_202792385 | 0.11 |
ENST00000549576.1
|
RP11-480I12.4
|
Putative inactive alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase-like protein LOC641515 |
| chr13_+_96329381 | 0.11 |
ENST00000602402.1
ENST00000376795.6 |
DNAJC3
|
DnaJ (Hsp40) homolog, subfamily C, member 3 |
| chr10_-_120938303 | 0.10 |
ENST00000356951.3
ENST00000298510.2 |
PRDX3
|
peroxiredoxin 3 |
| chr21_-_30365136 | 0.08 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr1_+_173446405 | 0.08 |
ENST00000340385.5
|
PRDX6
|
peroxiredoxin 6 |
| chr3_-_155524049 | 0.07 |
ENST00000534941.1
ENST00000340171.2 |
C3orf33
|
chromosome 3 open reading frame 33 |
| chr2_+_219524473 | 0.07 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr2_+_177025619 | 0.07 |
ENST00000410016.1
|
HOXD3
|
homeobox D3 |
| chr10_+_7860460 | 0.07 |
ENST00000344293.5
|
TAF3
|
TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa |
| chr13_-_37633567 | 0.06 |
ENST00000464744.1
|
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr10_+_50822083 | 0.05 |
ENST00000337653.2
ENST00000395562.2 |
CHAT
|
choline O-acetyltransferase |
| chr1_+_156611960 | 0.05 |
ENST00000361588.5
|
BCAN
|
brevican |
| chr19_+_57050317 | 0.05 |
ENST00000301318.3
ENST00000591844.1 |
ZFP28
|
ZFP28 zinc finger protein |
| chr8_-_144679296 | 0.05 |
ENST00000317198.6
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr15_-_43029252 | 0.05 |
ENST00000563260.1
ENST00000356231.3 |
CDAN1
|
codanin 1 |
| chr9_-_99616642 | 0.04 |
ENST00000478850.1
ENST00000481138.1 |
ZNF782
|
zinc finger protein 782 |
| chr1_+_156611704 | 0.04 |
ENST00000329117.5
|
BCAN
|
brevican |
| chr3_-_128902729 | 0.03 |
ENST00000451728.2
ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr1_+_22970119 | 0.03 |
ENST00000374640.4
ENST00000374639.3 ENST00000374637.1 |
C1QC
|
complement component 1, q subcomponent, C chain |
| chr2_+_219524379 | 0.03 |
ENST00000443791.1
ENST00000359273.3 ENST00000392109.1 ENST00000392110.2 ENST00000423377.1 ENST00000392111.2 ENST00000412366.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr19_+_5681011 | 0.03 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr19_-_17488143 | 0.01 |
ENST00000599426.1
ENST00000252590.4 |
PLVAP
|
plasmalemma vesicle associated protein |
| chr3_-_151176497 | 0.01 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr2_+_235860616 | 0.01 |
ENST00000392011.2
|
SH3BP4
|
SH3-domain binding protein 4 |
| chr20_-_54580523 | 0.00 |
ENST00000064571.2
|
CBLN4
|
cerebellin 4 precursor |
| chr17_-_74449252 | 0.00 |
ENST00000319380.7
|
UBE2O
|
ubiquitin-conjugating enzyme E2O |
| chr1_-_200379104 | 0.00 |
ENST00000367352.3
|
ZNF281
|
zinc finger protein 281 |
| chr13_+_50202435 | 0.00 |
ENST00000282026.1
|
ARL11
|
ADP-ribosylation factor-like 11 |
| chr22_-_50964558 | 0.00 |
ENST00000535425.1
ENST00000439934.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF350
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 1.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.3 | 0.8 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.2 | 1.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 1.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 4.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.5 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 1.2 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.1 | 3.9 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 3.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.3 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.4 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.2 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.2 | GO:1904796 | regulation of core promoter binding(GO:1904796) |
| 0.0 | 0.8 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.9 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 1.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 5.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 1.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 4.3 | GO:0005770 | late endosome(GO:0005770) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.3 | 5.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 0.7 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 1.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.4 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 1.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.3 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 4.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.8 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |