Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
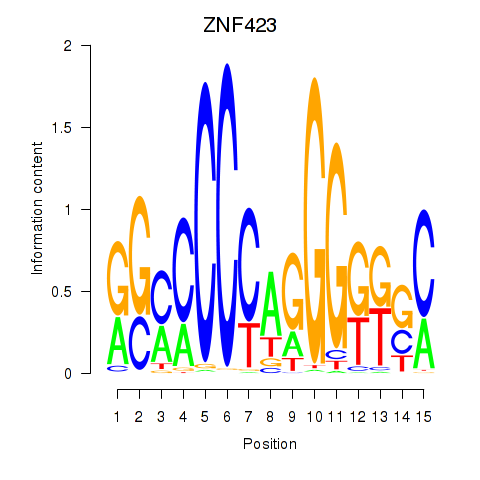
Results for ZNF423
Z-value: 1.00
Transcription factors associated with ZNF423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF423
|
ENSG00000102935.7 | zinc finger protein 423 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF423 | hg19_v2_chr16_-_49698136_49698209 | 0.50 | 5.4e-03 | Click! |
Activity profile of ZNF423 motif
Sorted Z-values of ZNF423 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_22022800 | 4.53 |
ENST00000397814.3
|
BMP1
|
bone morphogenetic protein 1 |
| chr5_+_150400124 | 4.42 |
ENST00000388825.4
ENST00000521650.1 ENST00000517973.1 |
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr6_+_151186554 | 3.40 |
ENST00000367321.3
ENST00000367307.4 |
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr11_-_61062762 | 3.24 |
ENST00000335613.5
|
VWCE
|
von Willebrand factor C and EGF domains |
| chr1_-_204121013 | 3.22 |
ENST00000367201.3
|
ETNK2
|
ethanolamine kinase 2 |
| chr1_-_204121102 | 3.21 |
ENST00000367202.4
|
ETNK2
|
ethanolamine kinase 2 |
| chr1_-_204121298 | 3.14 |
ENST00000367199.2
|
ETNK2
|
ethanolamine kinase 2 |
| chr19_-_11689752 | 3.14 |
ENST00000592659.1
ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr10_-_121296045 | 3.13 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr8_+_22022223 | 2.94 |
ENST00000306385.5
|
BMP1
|
bone morphogenetic protein 1 |
| chr5_+_135385202 | 2.55 |
ENST00000514554.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr8_+_22022653 | 2.49 |
ENST00000354870.5
ENST00000397816.3 ENST00000306349.8 |
BMP1
|
bone morphogenetic protein 1 |
| chr17_+_6347729 | 2.35 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr17_+_6347761 | 2.32 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr12_-_125348329 | 2.31 |
ENST00000546215.1
ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1
|
scavenger receptor class B, member 1 |
| chr16_+_71392616 | 2.28 |
ENST00000349553.5
ENST00000302628.4 ENST00000562305.1 |
CALB2
|
calbindin 2 |
| chr9_-_131486367 | 2.25 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr10_+_17271266 | 2.21 |
ENST00000224237.5
|
VIM
|
vimentin |
| chr22_-_37823468 | 2.18 |
ENST00000402918.2
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chrX_-_51239425 | 2.17 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr12_-_125348448 | 2.03 |
ENST00000339570.5
|
SCARB1
|
scavenger receptor class B, member 1 |
| chr11_+_60691924 | 2.00 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr1_-_155177677 | 1.69 |
ENST00000368378.3
ENST00000541990.1 ENST00000457183.2 |
THBS3
|
thrombospondin 3 |
| chr6_+_151187074 | 1.57 |
ENST00000367308.4
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chrX_-_107019181 | 1.56 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr10_+_135340859 | 1.51 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr20_-_62475273 | 1.50 |
ENST00000596861.1
|
AL158091.1
|
Protein LOC100509861 |
| chr15_+_41523417 | 1.50 |
ENST00000560397.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr11_-_61659006 | 1.48 |
ENST00000278829.2
|
FADS3
|
fatty acid desaturase 3 |
| chrX_+_49028265 | 1.47 |
ENST00000376322.3
ENST00000376327.5 |
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched) |
| chr7_-_23053719 | 1.47 |
ENST00000432176.2
ENST00000440481.1 |
FAM126A
|
family with sequence similarity 126, member A |
| chr7_-_23053693 | 1.47 |
ENST00000409763.1
ENST00000409923.1 |
FAM126A
|
family with sequence similarity 126, member A |
| chr10_-_76859247 | 1.46 |
ENST00000472493.2
ENST00000605915.1 ENST00000478873.2 |
DUSP13
|
dual specificity phosphatase 13 |
| chr3_-_48594248 | 1.43 |
ENST00000545984.1
ENST00000232375.3 ENST00000416568.1 ENST00000383734.2 ENST00000541519.1 ENST00000412035.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr11_-_61658853 | 1.40 |
ENST00000525588.1
ENST00000540820.1 |
FADS3
|
fatty acid desaturase 3 |
| chr17_+_7341586 | 1.36 |
ENST00000575235.1
|
FGF11
|
fibroblast growth factor 11 |
| chr9_+_112542572 | 1.34 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr8_+_22423219 | 1.29 |
ENST00000523965.1
ENST00000521554.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr19_-_10679644 | 1.27 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chrX_-_107018969 | 1.26 |
ENST00000372383.4
|
TSC22D3
|
TSC22 domain family, member 3 |
| chr9_+_116638562 | 1.26 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr16_+_27325202 | 1.17 |
ENST00000395762.2
ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R
|
interleukin 4 receptor |
| chr19_-_15311713 | 1.16 |
ENST00000601011.1
ENST00000263388.2 |
NOTCH3
|
notch 3 |
| chr19_+_45971246 | 1.05 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr17_+_36584662 | 1.05 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr2_+_62932779 | 1.04 |
ENST00000427809.1
ENST00000405482.1 ENST00000431489.1 |
EHBP1
|
EH domain binding protein 1 |
| chr8_+_22422749 | 1.02 |
ENST00000523900.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr3_-_195808980 | 1.01 |
ENST00000360110.4
|
TFRC
|
transferrin receptor |
| chr8_+_30241934 | 1.00 |
ENST00000538486.1
|
RBPMS
|
RNA binding protein with multiple splicing |
| chr10_+_124913793 | 0.98 |
ENST00000368865.4
ENST00000538238.1 ENST00000368859.2 |
BUB3
|
BUB3 mitotic checkpoint protein |
| chr3_-_195808952 | 0.97 |
ENST00000540528.1
ENST00000392396.3 ENST00000535031.1 ENST00000420415.1 |
TFRC
|
transferrin receptor |
| chr13_+_21277482 | 0.96 |
ENST00000304920.3
|
IL17D
|
interleukin 17D |
| chr1_+_155278625 | 0.95 |
ENST00000368356.4
ENST00000356657.6 |
FDPS
|
farnesyl diphosphate synthase |
| chr11_-_47207950 | 0.94 |
ENST00000298838.6
ENST00000531226.1 ENST00000524509.1 ENST00000528201.1 ENST00000530513.1 |
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr12_-_54779511 | 0.89 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr19_+_45582453 | 0.88 |
ENST00000591607.1
ENST00000591747.1 ENST00000270257.4 ENST00000391951.2 ENST00000587566.1 |
GEMIN7
MARK4
|
gem (nuclear organelle) associated protein 7 MAP/microtubule affinity-regulating kinase 4 |
| chr4_+_4388805 | 0.87 |
ENST00000504171.1
|
NSG1
|
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr6_+_33589161 | 0.83 |
ENST00000605930.1
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr11_+_61595752 | 0.80 |
ENST00000521849.1
|
FADS2
|
fatty acid desaturase 2 |
| chr6_+_142468361 | 0.78 |
ENST00000367630.4
|
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr5_-_172756506 | 0.77 |
ENST00000265087.4
|
STC2
|
stanniocalcin 2 |
| chr13_+_96743093 | 0.76 |
ENST00000376705.2
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chr22_+_35653445 | 0.73 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr4_-_20985632 | 0.73 |
ENST00000359001.5
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr9_-_35619539 | 0.71 |
ENST00000396757.1
|
CD72
|
CD72 molecule |
| chr3_+_6902794 | 0.69 |
ENST00000357716.4
ENST00000486284.1 ENST00000389336.4 ENST00000403881.1 ENST00000402647.2 |
GRM7
|
glutamate receptor, metabotropic 7 |
| chr17_-_48227877 | 0.69 |
ENST00000316878.6
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B |
| chr9_+_112542591 | 0.63 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr4_+_4388245 | 0.62 |
ENST00000433139.2
|
NSG1
|
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr1_+_55271736 | 0.62 |
ENST00000358193.3
ENST00000371273.3 |
C1orf177
|
chromosome 1 open reading frame 177 |
| chr6_-_35480640 | 0.61 |
ENST00000428978.1
ENST00000322263.4 |
TULP1
|
tubby like protein 1 |
| chr6_+_42896865 | 0.60 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr8_+_30241995 | 0.59 |
ENST00000397323.4
ENST00000339877.4 ENST00000320203.4 ENST00000287771.5 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr11_-_67120974 | 0.59 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr1_-_102462565 | 0.57 |
ENST00000370103.4
|
OLFM3
|
olfactomedin 3 |
| chr3_+_180630444 | 0.55 |
ENST00000491062.1
ENST00000468861.1 ENST00000445140.2 ENST00000484958.1 |
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chr11_-_64013663 | 0.53 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chrX_+_136648297 | 0.53 |
ENST00000287538.5
|
ZIC3
|
Zic family member 3 |
| chr6_-_35480705 | 0.52 |
ENST00000229771.6
|
TULP1
|
tubby like protein 1 |
| chr8_+_22423168 | 0.51 |
ENST00000518912.1
ENST00000428103.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr15_+_41523335 | 0.51 |
ENST00000334660.5
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr16_+_3014269 | 0.51 |
ENST00000575885.1
ENST00000571007.1 ENST00000319500.6 |
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr4_+_4387983 | 0.49 |
ENST00000397958.1
|
NSG1
|
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr12_-_118810688 | 0.49 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chr16_+_3014217 | 0.47 |
ENST00000572045.1
|
KREMEN2
|
kringle containing transmembrane protein 2 |
| chrX_+_106163626 | 0.46 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr12_-_50101003 | 0.45 |
ENST00000550488.1
|
FMNL3
|
formin-like 3 |
| chr6_-_91296602 | 0.43 |
ENST00000369325.3
ENST00000369327.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr18_-_60985914 | 0.42 |
ENST00000589955.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr13_-_46626847 | 0.42 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr19_+_20011775 | 0.40 |
ENST00000592245.1
ENST00000592160.1 ENST00000343769.5 |
AC007204.2
ZNF93
|
AC007204.2 zinc finger protein 93 |
| chr6_-_91296737 | 0.40 |
ENST00000369332.3
ENST00000369329.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr12_-_4554780 | 0.39 |
ENST00000228837.2
|
FGF6
|
fibroblast growth factor 6 |
| chr7_-_5570229 | 0.38 |
ENST00000331789.5
|
ACTB
|
actin, beta |
| chr4_+_128982490 | 0.37 |
ENST00000394288.3
ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr10_+_124914285 | 0.37 |
ENST00000407911.2
|
BUB3
|
BUB3 mitotic checkpoint protein |
| chr8_-_42234745 | 0.37 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr3_+_9958758 | 0.35 |
ENST00000383812.4
ENST00000438091.1 ENST00000295981.3 ENST00000436503.1 ENST00000403601.3 ENST00000416074.2 ENST00000455057.1 |
IL17RC
|
interleukin 17 receptor C |
| chr15_-_75660919 | 0.35 |
ENST00000569482.1
ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1
|
mannosidase, alpha, class 2C, member 1 |
| chr1_+_10459111 | 0.34 |
ENST00000541529.1
ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD
|
phosphogluconate dehydrogenase |
| chr11_-_62783276 | 0.33 |
ENST00000535878.1
ENST00000545207.1 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr2_+_171785824 | 0.33 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr7_+_20370746 | 0.30 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr7_-_100253993 | 0.30 |
ENST00000461605.1
ENST00000160382.5 |
ACTL6B
|
actin-like 6B |
| chr4_-_120548779 | 0.30 |
ENST00000264805.5
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr5_+_78407602 | 0.28 |
ENST00000274353.5
ENST00000524080.1 |
BHMT
|
betaine--homocysteine S-methyltransferase |
| chr7_-_27219849 | 0.27 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr9_+_135037334 | 0.27 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr1_+_53527854 | 0.27 |
ENST00000371500.3
ENST00000395871.2 ENST00000312553.5 |
PODN
|
podocan |
| chr5_-_176057365 | 0.27 |
ENST00000310112.3
|
SNCB
|
synuclein, beta |
| chr4_+_128982430 | 0.25 |
ENST00000512292.1
ENST00000508819.1 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr9_-_39239171 | 0.24 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr8_-_133493200 | 0.24 |
ENST00000388996.4
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3 |
| chr10_+_124913930 | 0.22 |
ENST00000368858.5
|
BUB3
|
BUB3 mitotic checkpoint protein |
| chr4_+_152330390 | 0.20 |
ENST00000503146.1
ENST00000435205.1 |
FAM160A1
|
family with sequence similarity 160, member A1 |
| chr2_+_171785012 | 0.20 |
ENST00000234160.4
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr5_-_176057518 | 0.20 |
ENST00000393693.2
|
SNCB
|
synuclein, beta |
| chr11_-_62783303 | 0.20 |
ENST00000336232.2
ENST00000430500.2 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr3_+_9958870 | 0.19 |
ENST00000413608.1
|
IL17RC
|
interleukin 17 receptor C |
| chrX_-_48827976 | 0.19 |
ENST00000218176.3
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr14_-_77608121 | 0.18 |
ENST00000319374.4
|
ZDHHC22
|
zinc finger, DHHC-type containing 22 |
| chr2_+_135011731 | 0.18 |
ENST00000281923.2
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr1_+_26644441 | 0.17 |
ENST00000374213.2
|
CD52
|
CD52 molecule |
| chr4_-_78740511 | 0.17 |
ENST00000504123.1
ENST00000264903.4 ENST00000515441.1 |
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like |
| chr7_+_150748288 | 0.16 |
ENST00000490540.1
|
ASIC3
|
acid-sensing (proton-gated) ion channel 3 |
| chr2_+_12246664 | 0.16 |
ENST00000449986.1
|
AC096559.1
|
AC096559.1 |
| chr7_-_44265492 | 0.15 |
ENST00000425809.1
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr10_-_75255724 | 0.15 |
ENST00000342558.3
ENST00000360663.5 ENST00000394829.2 ENST00000394828.2 ENST00000394822.2 |
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr3_+_196439275 | 0.14 |
ENST00000296333.5
|
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr22_-_38484922 | 0.14 |
ENST00000428572.1
|
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr9_-_16870704 | 0.10 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr16_-_11681023 | 0.09 |
ENST00000570904.1
ENST00000574701.1 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr19_-_57183114 | 0.09 |
ENST00000537055.2
ENST00000601659.1 |
ZNF835
|
zinc finger protein 835 |
| chr6_+_168841817 | 0.08 |
ENST00000356284.2
ENST00000354536.5 |
SMOC2
|
SPARC related modular calcium binding 2 |
| chr20_+_23016057 | 0.07 |
ENST00000255008.3
|
SSTR4
|
somatostatin receptor 4 |
| chr1_-_48462566 | 0.07 |
ENST00000606738.2
|
TRABD2B
|
TraB domain containing 2B |
| chr5_+_68788594 | 0.06 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chrX_+_30265256 | 0.06 |
ENST00000397548.2
|
MAGEB1
|
melanoma antigen family B, 1 |
| chr17_+_28705921 | 0.06 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr19_-_36391434 | 0.06 |
ENST00000396901.1
ENST00000585925.1 |
NFKBID
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta |
| chr6_+_30130969 | 0.05 |
ENST00000376694.4
|
TRIM15
|
tripartite motif containing 15 |
| chr3_+_196439170 | 0.05 |
ENST00000392391.3
ENST00000314118.4 |
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr12_+_7037461 | 0.03 |
ENST00000396684.2
|
ATN1
|
atrophin 1 |
| chr3_+_32726774 | 0.03 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr4_-_166034029 | 0.02 |
ENST00000306480.6
|
TMEM192
|
transmembrane protein 192 |
| chr14_-_93582214 | 0.02 |
ENST00000556603.2
ENST00000354313.3 |
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF423
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 1.2 | 5.0 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.7 | 4.4 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.6 | 3.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.4 | 10.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.4 | 2.2 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.4 | 9.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 2.8 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 1.5 | GO:0010193 | response to ozone(GO:0010193) |
| 0.3 | 2.0 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.2 | 1.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.2 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) response to odorant(GO:1990834) |
| 0.2 | 0.7 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.2 | 1.0 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.2 | 2.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.2 | 0.9 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 2.0 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.2 | 0.8 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.2 | 0.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.2 | GO:0072103 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 3.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 1.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 0.3 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 1.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 1.7 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 0.4 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.7 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.1 | 2.0 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 0.3 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.1 | 0.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.6 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 2.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.4 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 0.8 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.5 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 3.7 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.8 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 1.1 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 1.6 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 2.8 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.2 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:1904808 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.0 | 0.5 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 2.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 2.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.6 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.7 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 1.0 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.2 | 2.0 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.2 | 1.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 4.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 2.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.9 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 2.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 4.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 12.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 1.2 | 5.0 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 1.0 | 9.6 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.7 | 2.0 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.4 | 4.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.4 | 1.2 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.4 | 2.2 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.3 | 2.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 2.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 0.8 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 3.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 1.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 1.6 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 2.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 0.8 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.0 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 0.8 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.7 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 2.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 3.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.3 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.1 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 2.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.5 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 10.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 3.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 1.5 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 1.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.5 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 2.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 2.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 2.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 8.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 5.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 14.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.4 | 9.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 0.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 2.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 2.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 1.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 2.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |