Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
Results for ZNF652
Z-value: 0.72
Transcription factors associated with ZNF652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF652
|
ENSG00000198740.4 | zinc finger protein 652 |
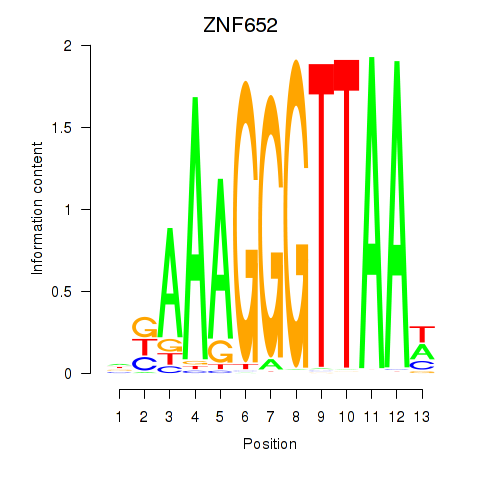
Activity profile of ZNF652 motif
Sorted Z-values of ZNF652 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_89224508 | 1.75 |
ENST00000525196.1
|
NOX4
|
NADPH oxidase 4 |
| chr11_-_89224488 | 1.43 |
ENST00000534731.1
ENST00000527626.1 |
NOX4
|
NADPH oxidase 4 |
| chr11_-_89224299 | 1.37 |
ENST00000343727.5
ENST00000531342.1 ENST00000375979.3 |
NOX4
|
NADPH oxidase 4 |
| chr7_+_80231466 | 1.30 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr22_-_37880543 | 1.15 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr7_+_23286182 | 1.09 |
ENST00000258733.4
ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB
|
glycoprotein (transmembrane) nmb |
| chr12_-_6982442 | 1.04 |
ENST00000523102.1
ENST00000524270.1 ENST00000519357.1 |
SPSB2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr10_-_28623368 | 1.04 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr11_-_89224638 | 1.02 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chr5_-_54281407 | 0.90 |
ENST00000381403.4
|
ESM1
|
endothelial cell-specific molecule 1 |
| chr16_-_56459354 | 0.85 |
ENST00000290649.5
|
AMFR
|
autocrine motility factor receptor, E3 ubiquitin protein ligase |
| chr11_-_111782484 | 0.84 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr5_+_135394840 | 0.73 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr11_-_111782696 | 0.69 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr17_+_48172639 | 0.68 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chr18_-_5296138 | 0.67 |
ENST00000400143.3
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr1_-_185597619 | 0.66 |
ENST00000608417.1
ENST00000436955.1 |
GS1-204I12.1
|
GS1-204I12.1 |
| chr9_+_74764278 | 0.63 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr1_-_6662919 | 0.61 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr5_+_167181917 | 0.61 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr2_+_38893208 | 0.59 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr16_-_79634595 | 0.56 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr17_+_72733350 | 0.56 |
ENST00000392613.5
ENST00000392612.3 ENST00000392610.1 |
RAB37
|
RAB37, member RAS oncogene family |
| chr17_-_6735012 | 0.55 |
ENST00000535086.1
|
TEKT1
|
tektin 1 |
| chr1_+_152635854 | 0.52 |
ENST00000368784.1
|
LCE2D
|
late cornified envelope 2D |
| chr19_+_46651474 | 0.52 |
ENST00000377693.4
|
IGFL2
|
IGF-like family member 2 |
| chr4_+_72897521 | 0.51 |
ENST00000308744.6
ENST00000344413.5 |
NPFFR2
|
neuropeptide FF receptor 2 |
| chr5_-_147211226 | 0.51 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr8_-_140715294 | 0.50 |
ENST00000303015.1
ENST00000520439.1 |
KCNK9
|
potassium channel, subfamily K, member 9 |
| chrX_-_128788914 | 0.49 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr12_+_6982725 | 0.49 |
ENST00000433346.1
|
LRRC23
|
leucine rich repeat containing 23 |
| chr20_-_36793663 | 0.48 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr19_-_14316980 | 0.46 |
ENST00000361434.3
ENST00000340736.6 |
LPHN1
|
latrophilin 1 |
| chrX_-_44202857 | 0.44 |
ENST00000420999.1
|
EFHC2
|
EF-hand domain (C-terminal) containing 2 |
| chr14_+_22771851 | 0.43 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39 |
| chr17_+_26800296 | 0.43 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr12_+_20848486 | 0.42 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr12_+_20848377 | 0.41 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr17_+_12569306 | 0.41 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr1_+_145293371 | 0.41 |
ENST00000342960.5
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr6_+_46655612 | 0.40 |
ENST00000544460.1
|
TDRD6
|
tudor domain containing 6 |
| chr17_+_26800648 | 0.39 |
ENST00000545060.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr11_-_63381925 | 0.39 |
ENST00000415826.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr3_+_121774202 | 0.39 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr17_+_26800756 | 0.39 |
ENST00000537681.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr10_+_104263743 | 0.38 |
ENST00000369902.3
ENST00000369899.2 ENST00000423559.2 |
SUFU
|
suppressor of fused homolog (Drosophila) |
| chr1_+_86934526 | 0.37 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr2_-_86564776 | 0.37 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr3_+_183353356 | 0.36 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr9_+_132096166 | 0.36 |
ENST00000436710.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr9_+_74764340 | 0.35 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr2_-_61244308 | 0.35 |
ENST00000407787.1
ENST00000398658.2 |
PUS10
|
pseudouridylate synthase 10 |
| chr13_+_111267866 | 0.34 |
ENST00000458711.2
ENST00000424185.2 ENST00000397191.4 ENST00000309957.2 |
CARKD
|
carbohydrate kinase domain containing |
| chr4_-_99578776 | 0.34 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr6_-_51952418 | 0.34 |
ENST00000371117.3
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr5_-_153857819 | 0.33 |
ENST00000231121.2
|
HAND1
|
heart and neural crest derivatives expressed 1 |
| chr4_-_165898768 | 0.33 |
ENST00000329314.5
|
TRIM61
|
tripartite motif containing 61 |
| chr1_+_202431859 | 0.33 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr11_-_86383370 | 0.33 |
ENST00000526834.1
ENST00000359636.2 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr14_-_88459503 | 0.33 |
ENST00000393568.4
ENST00000261304.2 |
GALC
|
galactosylceramidase |
| chr11_-_86383157 | 0.32 |
ENST00000393324.3
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr14_+_79745746 | 0.32 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr1_-_221915418 | 0.32 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr15_+_71185148 | 0.32 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr15_-_38856836 | 0.32 |
ENST00000450598.2
ENST00000559830.1 ENST00000558164.1 ENST00000310803.5 |
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr20_-_36793774 | 0.32 |
ENST00000361475.2
|
TGM2
|
transglutaminase 2 |
| chrX_-_110513703 | 0.32 |
ENST00000324068.1
|
CAPN6
|
calpain 6 |
| chr6_-_55740352 | 0.31 |
ENST00000370830.3
|
BMP5
|
bone morphogenetic protein 5 |
| chr14_+_79746249 | 0.30 |
ENST00000428277.2
|
NRXN3
|
neurexin 3 |
| chr15_-_49255632 | 0.30 |
ENST00000332408.4
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr17_-_4689727 | 0.29 |
ENST00000328739.5
ENST00000354194.4 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr16_+_31470179 | 0.29 |
ENST00000538189.1
ENST00000268314.4 |
ARMC5
|
armadillo repeat containing 5 |
| chr16_+_31470143 | 0.28 |
ENST00000457010.2
ENST00000563544.1 |
ARMC5
|
armadillo repeat containing 5 |
| chr14_+_24867992 | 0.28 |
ENST00000382554.3
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr7_+_1127723 | 0.28 |
ENST00000397088.3
|
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr16_-_57570450 | 0.28 |
ENST00000258214.2
|
CCDC102A
|
coiled-coil domain containing 102A |
| chr17_+_47865917 | 0.28 |
ENST00000259021.4
ENST00000454930.2 ENST00000509773.1 ENST00000510819.1 ENST00000424009.2 |
KAT7
|
K(lysine) acetyltransferase 7 |
| chr19_+_10531150 | 0.28 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr10_-_15413035 | 0.27 |
ENST00000378116.4
ENST00000455654.1 |
FAM171A1
|
family with sequence similarity 171, member A1 |
| chr3_-_133648656 | 0.27 |
ENST00000408895.2
|
C3orf36
|
chromosome 3 open reading frame 36 |
| chr4_-_77134742 | 0.27 |
ENST00000452464.2
|
SCARB2
|
scavenger receptor class B, member 2 |
| chr19_-_55881741 | 0.27 |
ENST00000264563.2
ENST00000590625.1 ENST00000585513.1 |
IL11
|
interleukin 11 |
| chr16_-_57513657 | 0.27 |
ENST00000566936.1
ENST00000568617.1 ENST00000567276.1 ENST00000569548.1 ENST00000569250.1 ENST00000564378.1 |
DOK4
|
docking protein 4 |
| chr14_-_22005343 | 0.26 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr12_+_32112340 | 0.26 |
ENST00000540924.1
ENST00000312561.4 |
KIAA1551
|
KIAA1551 |
| chr6_+_26217159 | 0.26 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr20_-_43753104 | 0.26 |
ENST00000372785.3
|
WFDC12
|
WAP four-disulfide core domain 12 |
| chrX_-_154250989 | 0.25 |
ENST00000360256.4
|
F8
|
coagulation factor VIII, procoagulant component |
| chr12_-_11139511 | 0.25 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr11_-_128894053 | 0.25 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr16_-_1843720 | 0.25 |
ENST00000415638.3
ENST00000215539.3 |
IGFALS
|
insulin-like growth factor binding protein, acid labile subunit |
| chr1_+_22351977 | 0.25 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr11_-_62752162 | 0.25 |
ENST00000458333.2
ENST00000421062.2 |
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr17_-_66287350 | 0.24 |
ENST00000580666.1
ENST00000583477.1 |
SLC16A6
|
solute carrier family 16, member 6 |
| chr2_+_11052054 | 0.24 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr14_-_65769392 | 0.24 |
ENST00000555736.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr3_+_49209023 | 0.24 |
ENST00000332780.2
|
KLHDC8B
|
kelch domain containing 8B |
| chr7_-_121944491 | 0.23 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr5_-_157002775 | 0.23 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr14_-_101034407 | 0.23 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr7_-_92465868 | 0.23 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr13_-_38172863 | 0.22 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr11_-_59951889 | 0.22 |
ENST00000532169.1
ENST00000534596.1 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr14_+_85996507 | 0.22 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr3_-_193272874 | 0.22 |
ENST00000342695.4
|
ATP13A4
|
ATPase type 13A4 |
| chr3_-_193272741 | 0.22 |
ENST00000392443.3
|
ATP13A4
|
ATPase type 13A4 |
| chr8_+_66955648 | 0.22 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr2_-_15701422 | 0.22 |
ENST00000441750.1
ENST00000281513.5 |
NBAS
|
neuroblastoma amplified sequence |
| chr14_-_69263043 | 0.22 |
ENST00000408913.2
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr11_+_57365150 | 0.22 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr1_+_22333943 | 0.21 |
ENST00000400271.2
|
CELA3A
|
chymotrypsin-like elastase family, member 3A |
| chr1_-_182641037 | 0.21 |
ENST00000483095.2
|
RGS8
|
regulator of G-protein signaling 8 |
| chr2_-_190044480 | 0.21 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr22_-_24641027 | 0.21 |
ENST00000398292.3
ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5
|
gamma-glutamyltransferase 5 |
| chr2_+_204193129 | 0.21 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr6_+_109169591 | 0.20 |
ENST00000368972.3
ENST00000392644.4 |
ARMC2
|
armadillo repeat containing 2 |
| chr2_-_192016316 | 0.20 |
ENST00000358470.4
ENST00000432798.1 ENST00000450994.1 |
STAT4
|
signal transducer and activator of transcription 4 |
| chr14_-_22005062 | 0.20 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr12_+_7072354 | 0.20 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr1_-_1182102 | 0.20 |
ENST00000330388.2
|
FAM132A
|
family with sequence similarity 132, member A |
| chr6_+_152126790 | 0.20 |
ENST00000456483.2
|
ESR1
|
estrogen receptor 1 |
| chr8_-_145018080 | 0.20 |
ENST00000354589.3
|
PLEC
|
plectin |
| chr7_-_127255982 | 0.19 |
ENST00000338516.3
ENST00000378740.2 |
PAX4
|
paired box 4 |
| chr1_-_156647189 | 0.19 |
ENST00000368223.3
|
NES
|
nestin |
| chr1_-_182640988 | 0.19 |
ENST00000367556.1
|
RGS8
|
regulator of G-protein signaling 8 |
| chr9_+_131683174 | 0.19 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr14_-_21562648 | 0.19 |
ENST00000555270.1
|
ZNF219
|
zinc finger protein 219 |
| chr12_+_10460549 | 0.19 |
ENST00000543420.1
ENST00000543777.1 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr19_+_45349630 | 0.19 |
ENST00000252483.5
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr12_-_6809958 | 0.19 |
ENST00000320591.5
ENST00000534837.1 |
PIANP
|
PILR alpha associated neural protein |
| chr15_+_68871569 | 0.19 |
ENST00000566799.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr5_-_78281775 | 0.19 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr5_-_178054014 | 0.19 |
ENST00000520957.1
|
CLK4
|
CDC-like kinase 4 |
| chr14_-_106926724 | 0.18 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr12_-_6451186 | 0.18 |
ENST00000540022.1
ENST00000536194.1 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr7_-_137686791 | 0.18 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr16_+_67927147 | 0.18 |
ENST00000291041.5
|
PSKH1
|
protein serine kinase H1 |
| chr1_+_181003067 | 0.17 |
ENST00000434571.2
ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1
|
major histocompatibility complex, class I-related |
| chr2_-_188378368 | 0.17 |
ENST00000392365.1
ENST00000435414.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr6_+_39760783 | 0.17 |
ENST00000398904.2
ENST00000538976.1 |
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr8_-_7309887 | 0.17 |
ENST00000458665.1
ENST00000528168.1 |
SPAG11B
|
sperm associated antigen 11B |
| chr14_-_21493884 | 0.17 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr11_-_26593649 | 0.17 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr3_+_46412345 | 0.17 |
ENST00000292303.4
|
CCR5
|
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr3_-_141719195 | 0.17 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr5_-_157002749 | 0.17 |
ENST00000517905.1
ENST00000430702.2 ENST00000394020.1 |
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr16_+_53242350 | 0.16 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr14_+_31343951 | 0.16 |
ENST00000556908.1
ENST00000555881.1 ENST00000460581.2 |
COCH
|
cochlin |
| chr15_+_63569785 | 0.16 |
ENST00000380343.4
ENST00000560353.1 |
APH1B
|
APH1B gamma secretase subunit |
| chr4_-_34041504 | 0.16 |
ENST00000512581.1
ENST00000505018.1 |
RP11-79E3.3
|
RP11-79E3.3 |
| chr12_-_6809543 | 0.16 |
ENST00000540656.1
|
PIANP
|
PILR alpha associated neural protein |
| chr16_-_75285380 | 0.16 |
ENST00000393420.6
ENST00000162330.5 |
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr11_-_59633951 | 0.16 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr20_-_60942361 | 0.15 |
ENST00000252999.3
|
LAMA5
|
laminin, alpha 5 |
| chr14_+_24407940 | 0.15 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr2_+_38893047 | 0.15 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr16_+_24266874 | 0.15 |
ENST00000005284.3
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr10_-_28571015 | 0.15 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr6_-_134373732 | 0.15 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr17_-_7120525 | 0.15 |
ENST00000447163.1
ENST00000399506.2 ENST00000302955.6 |
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr17_-_46035187 | 0.15 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr7_-_150675372 | 0.15 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr14_-_21493649 | 0.15 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr8_+_7716700 | 0.15 |
ENST00000454911.2
ENST00000326625.5 |
SPAG11A
|
sperm associated antigen 11A |
| chr6_-_28226984 | 0.14 |
ENST00000423974.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr3_+_135969148 | 0.14 |
ENST00000251654.4
ENST00000490504.1 ENST00000483687.1 ENST00000468777.1 ENST00000462637.1 ENST00000466072.1 ENST00000482086.1 ENST00000471595.1 ENST00000469217.1 ENST00000465423.1 ENST00000478469.1 |
PCCB
|
propionyl CoA carboxylase, beta polypeptide |
| chr4_-_143226979 | 0.14 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr9_+_109694914 | 0.14 |
ENST00000542028.1
|
ZNF462
|
zinc finger protein 462 |
| chr7_-_142583506 | 0.14 |
ENST00000359396.3
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr3_+_113616317 | 0.14 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr4_-_103998439 | 0.14 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr5_+_152870287 | 0.14 |
ENST00000340592.5
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr16_+_84733575 | 0.14 |
ENST00000219473.7
ENST00000563892.1 ENST00000562283.1 ENST00000570191.1 ENST00000569038.1 ENST00000570053.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr10_-_61513146 | 0.13 |
ENST00000430431.1
|
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr20_-_17662705 | 0.13 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr9_+_130860583 | 0.13 |
ENST00000373064.5
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr12_+_56546223 | 0.13 |
ENST00000550443.1
ENST00000207437.5 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr4_-_130014532 | 0.13 |
ENST00000506368.1
ENST00000439369.2 ENST00000503215.1 |
SCLT1
|
sodium channel and clathrin linker 1 |
| chr9_-_115983568 | 0.13 |
ENST00000446284.1
ENST00000414250.1 |
FKBP15
|
FK506 binding protein 15, 133kDa |
| chr9_+_130860810 | 0.13 |
ENST00000433501.1
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr1_-_33168336 | 0.13 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein |
| chr6_-_46048116 | 0.13 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr14_-_67982146 | 0.13 |
ENST00000557779.1
ENST00000557006.1 |
TMEM229B
|
transmembrane protein 229B |
| chr6_-_16761678 | 0.13 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr3_+_113251143 | 0.13 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr1_-_150849047 | 0.13 |
ENST00000354396.2
ENST00000505755.1 |
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr12_-_6451235 | 0.13 |
ENST00000440083.2
ENST00000162749.2 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr2_-_68547061 | 0.12 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr2_+_223536428 | 0.12 |
ENST00000446656.3
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1 |
| chr22_+_39966758 | 0.12 |
ENST00000407673.1
ENST00000401624.1 ENST00000404898.1 ENST00000402142.3 ENST00000336649.4 ENST00000400164.3 |
CACNA1I
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr16_-_30798492 | 0.12 |
ENST00000262525.4
|
ZNF629
|
zinc finger protein 629 |
| chr4_+_74275057 | 0.12 |
ENST00000511370.1
|
ALB
|
albumin |
| chr12_-_58329888 | 0.12 |
ENST00000546580.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr2_+_204193149 | 0.12 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr2_-_99224915 | 0.12 |
ENST00000328709.3
ENST00000409997.1 |
COA5
|
cytochrome c oxidase assembly factor 5 |
| chr3_+_151531810 | 0.12 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chr6_+_28227063 | 0.11 |
ENST00000343684.3
|
NKAPL
|
NFKB activating protein-like |
| chr17_-_41985096 | 0.11 |
ENST00000269095.4
ENST00000523220.1 |
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr15_-_34880646 | 0.11 |
ENST00000543376.1
|
GOLGA8A
|
golgin A8 family, member A |
| chr22_-_29196546 | 0.11 |
ENST00000403532.3
ENST00000216037.6 |
XBP1
|
X-box binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF652
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 1.0 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.2 | 0.7 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.2 | 1.3 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 0.7 | GO:2000724 | positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.2 | 5.6 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.2 | 0.5 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.4 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.1 | 1.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.4 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.9 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.3 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.3 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.1 | 0.3 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 0.5 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 1.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 1.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.2 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.1 | 0.3 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) regulation of lactation(GO:1903487) |
| 0.1 | 0.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.0 | 0.4 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.1 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.3 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.7 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.2 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.7 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.3 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.8 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.3 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.5 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.3 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.6 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.6 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.7 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.2 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.4 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.7 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 1.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.8 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.0 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.6 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.4 | 1.2 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.3 | 0.9 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.3 | 1.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 0.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.7 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.3 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 0.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.3 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.2 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.0 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 5.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |