Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
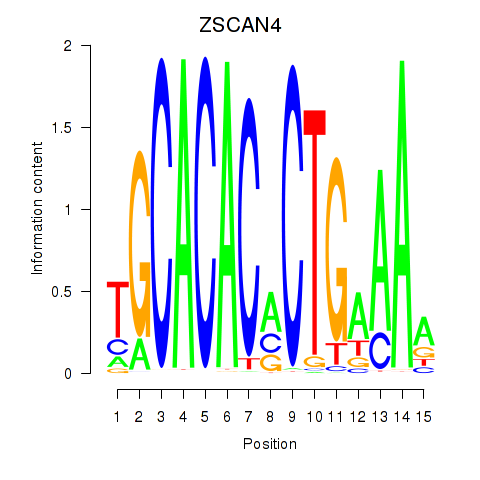
Results for ZSCAN4
Z-value: 0.63
Transcription factors associated with ZSCAN4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZSCAN4
|
ENSG00000180532.6 | zinc finger and SCAN domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZSCAN4 | hg19_v2_chr19_+_58180303_58180303 | 0.12 | 5.2e-01 | Click! |
Activity profile of ZSCAN4 motif
Sorted Z-values of ZSCAN4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_23712312 | 1.29 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr20_+_23168759 | 0.89 |
ENST00000411595.1
|
RP4-737E23.2
|
RP4-737E23.2 |
| chr4_+_74718906 | 0.72 |
ENST00000226524.3
|
PF4V1
|
platelet factor 4 variant 1 |
| chr12_-_57634475 | 0.72 |
ENST00000393825.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr12_-_10022735 | 0.66 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr12_-_96390063 | 0.66 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr5_+_31193847 | 0.60 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr2_-_101034070 | 0.54 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr7_-_84569561 | 0.54 |
ENST00000439105.1
|
AC074183.4
|
AC074183.4 |
| chr17_-_34207295 | 0.53 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr9_+_125137565 | 0.50 |
ENST00000373698.5
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr17_+_63133587 | 0.50 |
ENST00000449996.3
ENST00000262406.9 |
RGS9
|
regulator of G-protein signaling 9 |
| chr3_-_195538760 | 0.48 |
ENST00000475231.1
|
MUC4
|
mucin 4, cell surface associated |
| chr9_+_34652164 | 0.47 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr1_+_159409512 | 0.46 |
ENST00000423932.3
|
OR10J1
|
olfactory receptor, family 10, subfamily J, member 1 |
| chr17_-_46799872 | 0.45 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr9_+_128509624 | 0.45 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr15_+_40643227 | 0.45 |
ENST00000448599.2
|
PHGR1
|
proline/histidine/glycine-rich 1 |
| chr1_-_228603694 | 0.43 |
ENST00000366697.2
|
TRIM17
|
tripartite motif containing 17 |
| chr3_-_195538728 | 0.43 |
ENST00000349607.4
ENST00000346145.4 |
MUC4
|
mucin 4, cell surface associated |
| chr19_+_51630287 | 0.42 |
ENST00000599948.1
|
SIGLEC9
|
sialic acid binding Ig-like lectin 9 |
| chr5_-_82969405 | 0.41 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr14_-_55658252 | 0.41 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr17_+_9066252 | 0.40 |
ENST00000436734.1
|
NTN1
|
netrin 1 |
| chr15_+_75550940 | 0.40 |
ENST00000300576.5
|
GOLGA6C
|
golgin A6 family, member C |
| chr10_+_90354503 | 0.40 |
ENST00000531458.1
|
LIPJ
|
lipase, family member J |
| chr3_-_52569023 | 0.38 |
ENST00000307076.4
|
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr10_+_88414298 | 0.38 |
ENST00000372071.2
|
OPN4
|
opsin 4 |
| chr19_-_17516449 | 0.37 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr19_+_17516624 | 0.37 |
ENST00000596322.1
ENST00000600008.1 ENST00000601885.1 |
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr2_-_219696519 | 0.37 |
ENST00000545803.1
ENST00000430489.1 ENST00000392098.3 |
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr19_-_11266471 | 0.36 |
ENST00000592540.1
|
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr11_-_62783276 | 0.35 |
ENST00000535878.1
ENST00000545207.1 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr17_+_9728828 | 0.34 |
ENST00000262441.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr16_-_86542652 | 0.33 |
ENST00000599749.1
|
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr5_+_176784837 | 0.33 |
ENST00000408923.3
|
RGS14
|
regulator of G-protein signaling 14 |
| chr9_-_14910420 | 0.33 |
ENST00000380880.3
|
FREM1
|
FRAS1 related extracellular matrix 1 |
| chrX_-_20134990 | 0.33 |
ENST00000379651.3
ENST00000443379.3 ENST00000379643.5 |
MAP7D2
|
MAP7 domain containing 2 |
| chr22_-_24384260 | 0.32 |
ENST00000248935.5
|
GSTT1
|
glutathione S-transferase theta 1 |
| chr15_+_72947079 | 0.30 |
ENST00000421285.3
|
GOLGA6B
|
golgin A6 family, member B |
| chr8_+_85095769 | 0.30 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr15_-_74374891 | 0.29 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr2_-_219696511 | 0.29 |
ENST00000439262.2
|
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr15_-_28957469 | 0.29 |
ENST00000563027.1
ENST00000340249.3 |
GOLGA8M
|
golgin A8 family, member M |
| chr10_+_88414338 | 0.29 |
ENST00000241891.5
ENST00000443292.1 |
OPN4
|
opsin 4 |
| chr14_-_23904861 | 0.28 |
ENST00000355349.3
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta |
| chr19_+_30863271 | 0.28 |
ENST00000355537.3
|
ZNF536
|
zinc finger protein 536 |
| chr19_+_17516494 | 0.27 |
ENST00000534306.1
|
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr13_-_52733980 | 0.27 |
ENST00000339406.3
|
NEK3
|
NIMA-related kinase 3 |
| chr1_+_15764931 | 0.26 |
ENST00000375949.4
ENST00000375943.2 |
CTRC
|
chymotrypsin C (caldecrin) |
| chr19_-_9649253 | 0.26 |
ENST00000593003.1
|
ZNF426
|
zinc finger protein 426 |
| chr9_+_103189405 | 0.26 |
ENST00000395067.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr19_-_3801789 | 0.26 |
ENST00000590849.1
ENST00000395045.2 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr19_+_49436936 | 0.26 |
ENST00000221403.2
ENST00000523250.1 ENST00000522614.1 |
DHDH
|
dihydrodiol dehydrogenase (dimeric) |
| chr16_+_89228757 | 0.25 |
ENST00000565008.1
|
LINC00304
|
long intergenic non-protein coding RNA 304 |
| chr1_-_169337176 | 0.25 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr12_-_48164812 | 0.25 |
ENST00000549151.1
ENST00000548919.1 |
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr15_+_78632666 | 0.25 |
ENST00000299529.6
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr9_-_14910990 | 0.24 |
ENST00000380881.4
ENST00000422223.2 |
FREM1
|
FRAS1 related extracellular matrix 1 |
| chr8_+_26247878 | 0.24 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr17_+_2699697 | 0.24 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr8_+_85095497 | 0.23 |
ENST00000522455.1
ENST00000521695.1 |
RALYL
|
RALY RNA binding protein-like |
| chr3_+_98482175 | 0.23 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr10_+_18549645 | 0.22 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr15_+_75575176 | 0.22 |
ENST00000434739.3
|
GOLGA6D
|
golgin A6 family, member D |
| chr7_+_27282319 | 0.22 |
ENST00000222761.3
|
EVX1
|
even-skipped homeobox 1 |
| chr8_+_38831683 | 0.22 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr17_+_28256874 | 0.22 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr16_+_86229728 | 0.21 |
ENST00000601250.1
|
LINC01082
|
long intergenic non-protein coding RNA 1082 |
| chr4_-_152682129 | 0.21 |
ENST00000512306.1
ENST00000508611.1 ENST00000515812.1 ENST00000263985.6 |
PET112
|
PET112 homolog (yeast) |
| chr14_-_106610852 | 0.21 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr19_+_17516531 | 0.21 |
ENST00000528911.1
ENST00000528604.1 ENST00000595892.1 ENST00000500836.2 ENST00000598546.1 ENST00000600369.1 ENST00000598356.1 ENST00000594426.1 |
MVB12A
CTD-2521M24.9
|
multivesicular body subunit 12A CTD-2521M24.9 |
| chr8_-_72987810 | 0.20 |
ENST00000262209.4
|
TRPA1
|
transient receptor potential cation channel, subfamily A, member 1 |
| chr21_+_46066331 | 0.20 |
ENST00000334670.8
|
KRTAP10-11
|
keratin associated protein 10-11 |
| chr16_+_16481306 | 0.20 |
ENST00000422673.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr13_-_114898016 | 0.19 |
ENST00000542651.1
ENST00000334062.7 |
RASA3
|
RAS p21 protein activator 3 |
| chr4_-_168155300 | 0.19 |
ENST00000541637.1
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr1_+_89829610 | 0.19 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr15_+_38544476 | 0.18 |
ENST00000299084.4
|
SPRED1
|
sprouty-related, EVH1 domain containing 1 |
| chr2_-_36779411 | 0.18 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chrX_-_62571220 | 0.18 |
ENST00000374884.2
|
SPIN4
|
spindlin family, member 4 |
| chr16_+_6069664 | 0.18 |
ENST00000422070.4
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr8_-_16050214 | 0.17 |
ENST00000262101.5
|
MSR1
|
macrophage scavenger receptor 1 |
| chrX_-_62571187 | 0.17 |
ENST00000335144.3
|
SPIN4
|
spindlin family, member 4 |
| chr8_-_16050288 | 0.17 |
ENST00000350896.3
|
MSR1
|
macrophage scavenger receptor 1 |
| chr4_-_168155169 | 0.16 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr17_+_58499844 | 0.16 |
ENST00000269127.4
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr21_+_45879814 | 0.16 |
ENST00000596691.1
|
LRRC3DN
|
LRRC3 downstream neighbor (non-protein coding) |
| chr2_+_240323439 | 0.15 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr3_-_53290016 | 0.15 |
ENST00000423525.2
ENST00000423516.1 ENST00000296289.6 ENST00000462138.1 |
TKT
|
transketolase |
| chr3_+_52009110 | 0.14 |
ENST00000491470.1
|
ABHD14A
|
abhydrolase domain containing 14A |
| chr1_+_157963063 | 0.14 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr5_+_149887672 | 0.14 |
ENST00000261797.6
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr4_-_168155417 | 0.14 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr6_-_153304697 | 0.14 |
ENST00000367241.3
|
FBXO5
|
F-box protein 5 |
| chr6_-_76203454 | 0.14 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr3_+_9944303 | 0.13 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr6_-_34113856 | 0.13 |
ENST00000538487.2
|
GRM4
|
glutamate receptor, metabotropic 4 |
| chr12_-_96390108 | 0.13 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr6_-_76203345 | 0.13 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr16_+_15737124 | 0.13 |
ENST00000396355.1
ENST00000396353.2 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr7_+_107224364 | 0.13 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr16_+_16472912 | 0.13 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr7_-_99381884 | 0.13 |
ENST00000336411.2
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chrX_+_2984874 | 0.13 |
ENST00000359361.2
|
ARSF
|
arylsulfatase F |
| chr21_-_33651324 | 0.13 |
ENST00000290130.3
|
MIS18A
|
MIS18 kinetochore protein A |
| chr3_+_9944492 | 0.13 |
ENST00000383814.3
ENST00000454190.2 ENST00000454992.1 |
IL17RE
|
interleukin 17 receptor E |
| chr3_+_14716606 | 0.12 |
ENST00000253697.3
ENST00000435614.1 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr1_+_226736446 | 0.12 |
ENST00000366788.3
ENST00000366789.4 |
C1orf95
|
chromosome 1 open reading frame 95 |
| chr18_+_6729698 | 0.12 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr6_+_29795595 | 0.12 |
ENST00000360323.6
ENST00000376818.3 ENST00000376815.3 |
HLA-G
|
major histocompatibility complex, class I, G |
| chr20_-_60573188 | 0.12 |
ENST00000474089.1
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chr3_+_130569592 | 0.11 |
ENST00000533801.2
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr15_+_63413990 | 0.11 |
ENST00000261893.4
|
LACTB
|
lactamase, beta |
| chr11_+_45792967 | 0.11 |
ENST00000378779.2
|
CTD-2210P24.4
|
Uncharacterized protein |
| chr21_+_25801041 | 0.11 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr13_+_28813645 | 0.11 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr17_-_58499698 | 0.11 |
ENST00000590297.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr3_+_111393501 | 0.11 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr10_+_82009466 | 0.11 |
ENST00000356374.4
|
AL359195.1
|
Uncharacterized protein; cDNA FLJ46261 fis, clone TESTI4025062 |
| chr15_-_41836441 | 0.10 |
ENST00000567866.1
ENST00000561603.1 ENST00000304330.4 ENST00000566863.1 |
RPAP1
|
RNA polymerase II associated protein 1 |
| chr7_-_99381798 | 0.10 |
ENST00000415003.1
ENST00000354593.2 |
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr3_+_50284321 | 0.10 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr4_+_15779901 | 0.10 |
ENST00000226279.3
|
CD38
|
CD38 molecule |
| chr16_-_86542455 | 0.10 |
ENST00000595886.1
ENST00000597578.1 ENST00000593604.1 |
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chrX_+_1387693 | 0.10 |
ENST00000381529.3
ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr4_-_168155730 | 0.10 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr1_-_110283138 | 0.10 |
ENST00000256594.3
|
GSTM3
|
glutathione S-transferase mu 3 (brain) |
| chr4_-_168155577 | 0.10 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_-_168155700 | 0.10 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr19_+_571277 | 0.09 |
ENST00000346916.4
ENST00000545507.2 |
BSG
|
basigin (Ok blood group) |
| chr1_-_161600822 | 0.09 |
ENST00000534776.1
ENST00000540048.1 |
FCGR3B
FCGR3A
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chrX_+_17393543 | 0.09 |
ENST00000380060.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr12_+_16064106 | 0.09 |
ENST00000428559.2
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr11_-_128812744 | 0.09 |
ENST00000458238.2
ENST00000531399.1 ENST00000602346.1 |
TP53AIP1
|
tumor protein p53 regulated apoptosis inducing protein 1 |
| chr12_+_66218212 | 0.08 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chrX_+_56259316 | 0.08 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr1_-_177133818 | 0.07 |
ENST00000424564.2
ENST00000361833.2 |
ASTN1
|
astrotactin 1 |
| chr15_+_63414017 | 0.07 |
ENST00000413507.2
|
LACTB
|
lactamase, beta |
| chr13_+_60971080 | 0.07 |
ENST00000377894.2
|
TDRD3
|
tudor domain containing 3 |
| chr2_+_234545092 | 0.07 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr2_+_220436917 | 0.07 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr1_+_169337172 | 0.07 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr8_-_16050142 | 0.07 |
ENST00000536385.1
ENST00000381998.4 |
MSR1
|
macrophage scavenger receptor 1 |
| chr6_-_32083106 | 0.07 |
ENST00000442721.1
|
TNXB
|
tenascin XB |
| chr1_+_209929377 | 0.07 |
ENST00000400959.3
ENST00000367025.3 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr17_-_73761222 | 0.07 |
ENST00000437911.1
ENST00000225614.2 |
GALK1
|
galactokinase 1 |
| chr15_-_62457480 | 0.06 |
ENST00000380392.3
|
C2CD4B
|
C2 calcium-dependent domain containing 4B |
| chr19_-_42636617 | 0.06 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr17_-_7080227 | 0.06 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr11_+_111750206 | 0.06 |
ENST00000530214.1
ENST00000530799.1 |
C11orf1
|
chromosome 11 open reading frame 1 |
| chrX_+_1387721 | 0.06 |
ENST00000419094.1
ENST00000381509.3 ENST00000494969.2 ENST00000355805.2 ENST00000355432.3 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr12_-_11548496 | 0.06 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chrX_+_103294483 | 0.05 |
ENST00000355016.3
|
H2BFM
|
H2B histone family, member M |
| chr1_+_231376941 | 0.05 |
ENST00000436239.1
ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr2_-_167232484 | 0.05 |
ENST00000375387.4
ENST00000303354.6 ENST00000409672.1 |
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit |
| chr16_-_80838195 | 0.05 |
ENST00000570137.2
|
CDYL2
|
chromodomain protein, Y-like 2 |
| chr9_+_139557360 | 0.05 |
ENST00000308874.7
ENST00000406555.3 ENST00000492862.2 |
EGFL7
|
EGF-like-domain, multiple 7 |
| chr12_+_41086215 | 0.05 |
ENST00000547702.1
ENST00000551424.1 |
CNTN1
|
contactin 1 |
| chr5_+_61602055 | 0.05 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr22_+_18121356 | 0.05 |
ENST00000317582.5
ENST00000543133.1 ENST00000538149.1 ENST00000337612.5 ENST00000493680.1 |
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr12_+_133613878 | 0.05 |
ENST00000392319.2
ENST00000543758.1 |
ZNF84
|
zinc finger protein 84 |
| chr3_-_58572760 | 0.05 |
ENST00000447756.2
|
FAM107A
|
family with sequence similarity 107, member A |
| chr1_-_153588765 | 0.04 |
ENST00000368701.1
ENST00000344616.2 |
S100A14
|
S100 calcium binding protein A14 |
| chr18_+_32173276 | 0.04 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr12_+_16064258 | 0.04 |
ENST00000524480.1
ENST00000531803.1 ENST00000532964.1 |
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr1_+_2036149 | 0.04 |
ENST00000482686.1
ENST00000400920.1 ENST00000486681.1 |
PRKCZ
|
protein kinase C, zeta |
| chr19_-_35626104 | 0.04 |
ENST00000310123.3
ENST00000392225.3 |
LGI4
|
leucine-rich repeat LGI family, member 4 |
| chr13_+_60971427 | 0.04 |
ENST00000535286.1
ENST00000377881.2 |
TDRD3
|
tudor domain containing 3 |
| chr14_+_53173890 | 0.04 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr1_-_231376867 | 0.04 |
ENST00000366649.2
ENST00000318906.2 ENST00000366651.3 |
C1orf131
|
chromosome 1 open reading frame 131 |
| chr14_+_53173910 | 0.04 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr15_+_76135622 | 0.03 |
ENST00000338677.4
ENST00000267938.4 ENST00000569423.1 |
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr16_+_23847267 | 0.03 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr3_+_130569429 | 0.03 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr17_-_67057203 | 0.03 |
ENST00000340001.4
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr5_-_172036436 | 0.03 |
ENST00000601856.1
|
AC027309.1
|
AC027309.1 |
| chr6_+_114178512 | 0.03 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr9_-_114090713 | 0.03 |
ENST00000302681.1
ENST00000374428.1 |
OR2K2
|
olfactory receptor, family 2, subfamily K, member 2 |
| chr6_-_42690312 | 0.02 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr12_+_122242597 | 0.02 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr21_-_46221684 | 0.02 |
ENST00000330942.5
|
UBE2G2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr11_+_61717535 | 0.02 |
ENST00000534553.1
ENST00000301774.9 |
BEST1
|
bestrophin 1 |
| chr3_+_151531810 | 0.02 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chr1_+_52195542 | 0.02 |
ENST00000462759.1
ENST00000486942.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr7_-_135194822 | 0.01 |
ENST00000428680.2
ENST00000315544.5 ENST00000423368.2 ENST00000451834.1 ENST00000361528.4 ENST00000356162.4 ENST00000541284.1 |
CNOT4
|
CCR4-NOT transcription complex, subunit 4 |
| chr11_+_125462690 | 0.01 |
ENST00000392708.4
ENST00000529196.1 ENST00000531491.1 |
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chrX_+_119495934 | 0.01 |
ENST00000218008.3
ENST00000361319.3 ENST00000539306.1 |
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr17_+_30348024 | 0.01 |
ENST00000327564.7
ENST00000584368.1 ENST00000394713.3 ENST00000341671.7 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr19_-_42636543 | 0.01 |
ENST00000528894.4
ENST00000560804.2 ENST00000560558.1 ENST00000560398.1 ENST00000526816.2 |
POU2F2
|
POU class 2 homeobox 2 |
| chr1_-_161519682 | 0.01 |
ENST00000367969.3
ENST00000443193.1 |
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr19_-_10530784 | 0.01 |
ENST00000593124.1
|
CDC37
|
cell division cycle 37 |
| chr1_-_171621815 | 0.01 |
ENST00000037502.6
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr1_-_161519579 | 0.00 |
ENST00000426740.1
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr7_-_155326532 | 0.00 |
ENST00000406197.1
ENST00000321736.5 |
CNPY1
|
canopy FGF signaling regulator 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZSCAN4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.4 | GO:1901253 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 1.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.3 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.3 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 0.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.4 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.3 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 0.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.2 | GO:0036378 | alkaloid catabolic process(GO:0009822) calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.1 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0050968 | thermoception(GO:0050955) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.7 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.3 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0031049 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0015014 | protein sulfation(GO:0006477) heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.0 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.0 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 1.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.8 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.3 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.7 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.3 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.2 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.0 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 1.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.0 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |