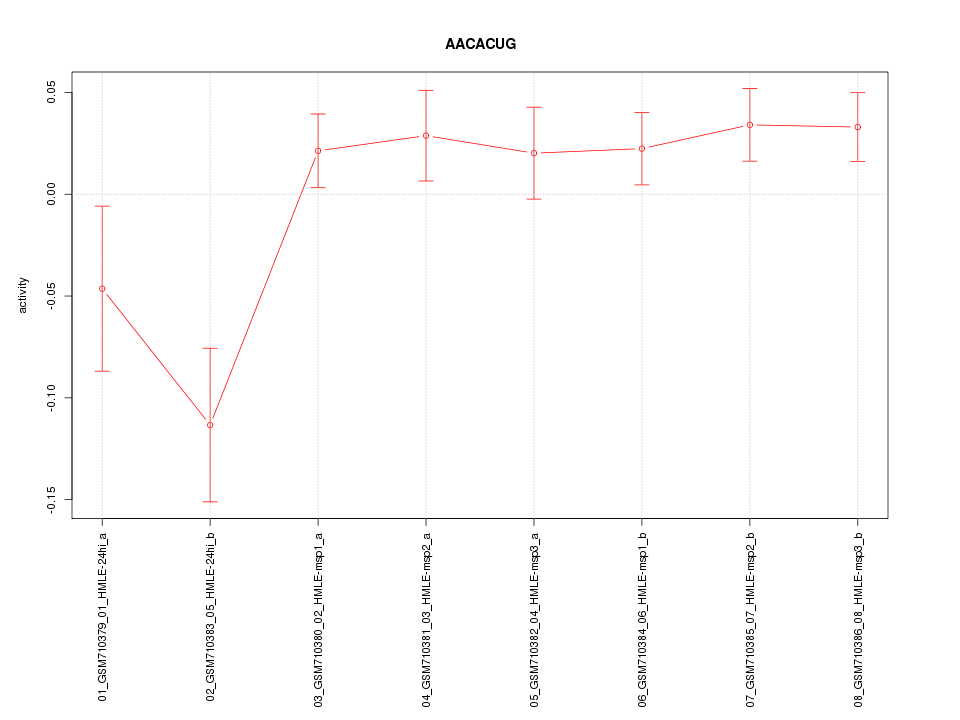
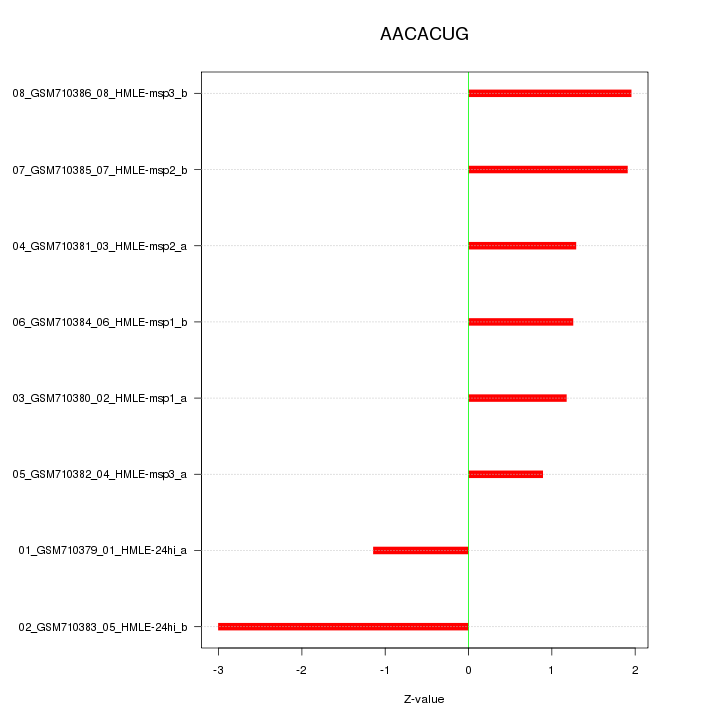
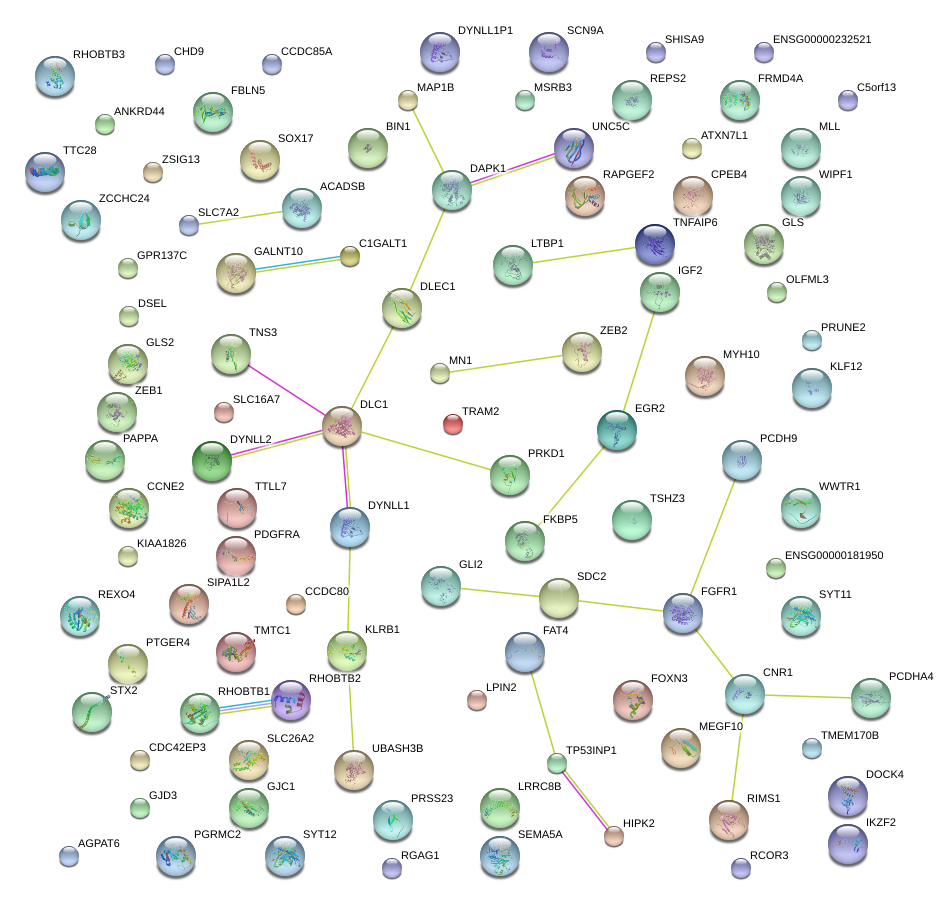
|
chr10_+_31608097
|
2.572
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr10_+_31610063
|
2.384
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr8_-_12973752
|
2.134
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr8_-_12990731
|
2.067
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr2_-_145277879
|
1.916
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr6_+_72926462
|
1.748
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_+_72922513
|
1.688
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_-_52441815
|
1.630
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr2_-_214016332
|
1.581
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr6_+_72926144
|
1.579
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr7_-_47621741
|
1.487
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr6_+_73076431
|
1.432
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr8_+_97505876
|
1.396
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr10_-_81205091
|
1.382
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr5_+_95066628
|
1.342
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr4_+_55095263
|
1.187
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr9_+_90112754
|
1.145
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr22_-_29075708
|
1.118
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr12_-_29936685
|
1.051
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr13_-_67804432
|
1.022
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr2_-_214015053
|
1.002
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr1_+_89990396
|
0.997
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr12_-_29937691
|
0.996
|
NM_175861
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr6_+_11538486
|
0.985
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr14_-_89883306
|
0.978
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr11_-_2162340
|
0.971
|
NM_001127598
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr14_-_92414004
|
0.934
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr14_-_30396811
|
0.931
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr11_-_2170792
|
0.911
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr11_+_122526339
|
0.908
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr8_+_55370494
|
0.899
|
NM_022454
|
SOX17
|
SRY (sex determining region Y)-box 17
|
|
chr7_-_105319608
|
0.869
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr3_-_112359962
|
0.861
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_90024271
|
0.858
|
NM_015350
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr12_-_131323616
|
0.845
|
NM_001980
NM_194356
|
STX2
|
syntaxin 2
|
|
chr8_-_38325241
|
0.843
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr11_-_2160199
|
0.830
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr10_-_14372807
|
0.828
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr8_-_38326351
|
0.826
|
NM_001174063
NM_001174064
NM_015850
NM_023105
NM_023106
NM_023110
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr14_-_90085325
|
0.810
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr6_-_88875766
|
0.776
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr6_+_72596648
|
0.770
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr5_-_9546157
|
0.759
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr6_-_35696359
|
0.758
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr4_-_129209983
|
0.734
|
NM_006320
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr18_-_3011944
|
0.723
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr8_-_95907423
|
0.710
|
NM_057749
|
CCNE2
|
cyclin E2
|
|
chr2_+_56411123
|
0.705
|
NM_001080433
|
CCDC85A
|
coiled-coil domain containing 85A
|
|
chr1_-_232651227
|
0.691
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr5_+_173315313
|
0.682
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr7_-_111846384
|
0.681
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr22_-_28197469
|
0.665
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr12_+_60083125
|
0.662
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr1_-_84464728
|
0.656
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr9_-_79520878
|
0.650
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr5_+_71403040
|
0.648
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr2_+_33172368
|
0.646
|
NM_206943
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr4_+_160188997
|
0.642
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr2_+_33359607
|
0.642
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr8_+_17354562
|
0.635
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr8_-_95961543
|
0.626
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr2_+_191745546
|
0.626
|
NM_014905
|
GLS
|
glutaminase
|
|
chr17_-_8534008
|
0.616
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr2_-_175499275
|
0.615
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr2_-_175547471
|
0.612
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr18_-_65183922
|
0.612
|
NM_032160
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr1_+_211433285
|
0.611
|
NM_018254
|
RCOR3
|
REST corepressor 3
|
|
chr19_-_31840118
|
0.593
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr2_+_152214105
|
0.592
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr5_+_149340287
|
0.589
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr1_+_114522029
|
0.587
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr6_-_35656677
|
0.580
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr2_-_198175494
|
0.570
|
NM_001195144
NM_153697
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr16_+_12995454
|
0.558
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr2_-_37899227
|
0.551
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr11_+_86511490
|
0.550
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr1_+_155829259
|
0.549
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr5_-_111093251
|
0.536
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr9_+_118916069
|
0.531
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr5_-_111093030
|
0.529
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111312522
|
0.526
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr11_-_105892908
|
0.525
|
NM_032424
|
KIAA1826
|
KIAA1826
|
|
chr5_-_111092918
|
0.524
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chrX_+_16964730
|
0.520
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr5_-_111093792
|
0.515
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr8_+_41435666
|
0.513
|
NM_178819
|
AGPAT6
|
1-acylglycerol-3-phosphate O-acyltransferase 6 (lysophosphatidic acid acyltransferase, zeta)
|
|
chr16_+_53088791
|
0.513
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr2_-_127864505
|
0.511
|
NM_004305
NM_139343
NM_139344
NM_139345
NM_139346
NM_139347
NM_139348
NM_139349
NM_139350
NM_139351
|
BIN1
|
bridging integrator 1
|
|
chr11_+_118307072
|
0.510
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr17_-_42908178
|
0.508
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr12_+_65672422
|
0.506
|
NM_001031679
NM_001193460
NM_198080
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr4_-_96470350
|
0.504
|
NM_003728
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr12_+_65672682
|
0.502
|
NM_001193461
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr8_+_17396285
|
0.499
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr10_+_124768401
|
0.492
|
NM_001609
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chr2_-_167232464
|
0.491
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr4_+_126237566
|
0.488
|
NM_024582
|
FAT4
|
FAT tumor suppressor homolog 4 (Drosophila)
|
|
chr8_-_13372183
|
0.482
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr17_-_42907606
|
0.479
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chrX_+_109662284
|
0.472
|
NM_020769
|
RGAG1
|
retrotransposon gag domain containing 1
|
|
chr5_+_153570270
|
0.462
|
NM_198321
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10)
|
|
chr2_+_121554866
|
0.459
|
NM_005270
|
GLI2
|
GLI family zinc finger 2
|
|
chr7_-_139477437
|
0.455
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr7_+_7222142
|
0.454
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr5_+_40680021
|
0.452
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr14_+_53019865
|
0.447
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr10_-_62703822
|
0.444
|
NM_001242359
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr10_-_64578926
|
0.438
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr5_+_126626455
|
0.437
|
NM_032446
|
MEGF10
|
multiple EGF-like-domains 10
|
|
chr3_-_149375584
|
0.437
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr13_-_74707913
|
0.429
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr17_+_61699786
|
0.429
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chrX_+_100333835
|
0.428
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr16_-_20911560
|
0.427
|
NM_173475
|
DCUN1D3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr8_-_57123858
|
0.424
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr15_-_49338496
|
0.422
|
NM_001193489
NM_014701
|
SECISBP2L
|
SECIS binding protein 2-like
|
|
chr1_+_159141376
|
0.421
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chrX_-_99665270
|
0.418
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr17_+_47074749
|
0.416
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr17_+_68165660
|
0.415
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chrX_-_99986496
|
0.414
|
NM_001174068
NM_080737
|
SYTL4
|
synaptotagmin-like 4
|
|
chr5_-_149535420
|
0.409
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr1_+_211432693
|
0.408
|
NM_001136223
NM_001136224
NM_001136225
|
RCOR3
|
REST corepressor 3
|
|
chr8_+_17433941
|
0.405
|
NM_006207
|
PDGFRL
|
platelet-derived growth factor receptor-like
|
|
chr3_+_187930720
|
0.402
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr5_-_41510627
|
0.399
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr4_-_57522469
|
0.399
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chrX_-_99987072
|
0.398
|
NM_001129896
|
SYTL4
|
synaptotagmin-like 4
|
|
chr2_+_201450730
|
0.397
|
NM_001159
|
AOX1
|
aldehyde oxidase 1
|
|
chr5_+_72143929
|
0.396
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr13_+_32605436
|
0.395
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr22_-_44708730
|
0.393
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr9_-_6015606
|
0.393
|
NM_001243202
NM_001243203
NM_012416
|
RANBP6
|
RAN binding protein 6
|
|
chr17_-_78450247
|
0.392
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr11_-_74109417
|
0.391
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr12_-_108714438
|
0.388
|
NM_001142345
|
CMKLR1
|
chemokine-like receptor 1
|
|
chr10_-_33623771
|
0.386
|
NM_001024628
NM_001024629
NM_001244972
NM_001244973
NM_003873
|
NRP1
|
neuropilin 1
|
|
chr15_+_63569748
|
0.386
|
NM_001145646
NM_031301
|
APH1B
|
anterior pharynx defective 1 homolog B (C. elegans)
|
|
chr7_-_100286869
|
0.382
|
NM_022574
|
GIGYF1
|
GRB10 interacting GYF protein 1
|
|
chr16_+_31191430
|
0.379
|
NM_001170634
NM_001170937
NM_004960
|
FUS
|
fused in sarcoma
|
|
chr3_+_63850181
|
0.378
|
NM_000333
|
ATXN7
|
ataxin 7
|
|
chr11_-_33744272
|
0.373
|
NM_001127223
NM_001127225
NM_001127226
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr12_+_26126687
|
0.372
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr1_-_33168356
|
0.371
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr10_-_64576123
|
0.369
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr12_-_108733075
|
0.368
|
NM_001142343
NM_001142344
NM_004072
|
CMKLR1
|
chemokine-like receptor 1
|
|
chr3_+_187871473
|
0.365
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr2_-_218808705
|
0.363
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr8_-_122653493
|
0.362
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr4_-_57547845
|
0.361
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr3_-_16555128
|
0.356
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr9_-_89562103
|
0.350
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr11_-_33744497
|
0.350
|
NM_001127227
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr3_+_69812620
|
0.348
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr2_-_163099877
|
0.348
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr3_+_69788562
|
0.348
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_69812961
|
0.346
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_69915374
|
0.345
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr10_+_76586299
|
0.341
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr1_+_118148506
|
0.341
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr14_-_61115906
|
0.339
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr5_+_172068188
|
0.337
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr5_-_172198160
|
0.336
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr13_-_77460432
|
0.336
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr12_+_12938540
|
0.335
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr2_-_225907312
|
0.335
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr3_+_154797644
|
0.334
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr3_-_149421059
|
0.334
|
NM_001168278
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr3_+_154797435
|
0.333
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr3_-_114343791
|
0.332
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_-_114102488
|
0.331
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr13_+_93879011
|
0.330
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr14_+_103801160
|
0.328
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr13_-_110438896
|
0.328
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr5_-_121412805
|
0.327
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr6_-_88854989
|
0.327
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr9_-_14693459
|
0.326
|
NM_178566
|
ZDHHC21
|
zinc finger, DHHC-type containing 21
|
|
chr15_-_60919642
|
0.326
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr9_+_4490193
|
0.325
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr9_+_32384560
|
0.324
|
NM_002197
|
ACO1
|
aconitase 1, soluble
|
|
chr3_+_154797910
|
0.323
|
NM_007288
|
MME
|
membrane metallo-endopeptidase
|
|
chr11_-_63536112
|
0.320
|
NM_001144936
|
C11orf95
|
chromosome 11 open reading frame 95
|
|
chr5_-_121414011
|
0.320
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr3_-_8811263
|
0.317
|
NM_000916
|
OXTR
|
oxytocin receptor
|
|
chr8_+_16884745
|
0.316
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr1_+_245133628
|
0.316
|
NM_001143943
|
EFCAB2
|
EF-hand calcium binding domain 2
|
|
chr3_+_154798078
|
0.316
|
NM_007289
|
MME
|
membrane metallo-endopeptidase
|
|
chr17_-_58603492
|
0.316
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr2_+_46926323
|
0.313
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr17_+_76000248
|
0.312
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr10_+_116581495
|
0.312
|
NM_001135051
NM_020940
|
FAM160B1
|
family with sequence similarity 160, member B1
|
|
chr14_+_50999799
|
0.310
|
NM_001127713
|
ATL1
|
atlastin GTPase 1
|
|
chr5_-_111091947
|
0.310
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_-_178789597
|
0.310
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr1_-_225840660
|
0.308
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr17_-_49198084
|
0.308
|
NM_001130527
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr5_+_72112417
|
0.306
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr11_-_33757872
|
0.305
|
NM_000611
NM_203329
NM_203330
NM_203331
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr4_-_138453628
|
0.302
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr5_-_95158465
|
0.301
|
NM_001118890
NM_001243658
NM_001243659
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|