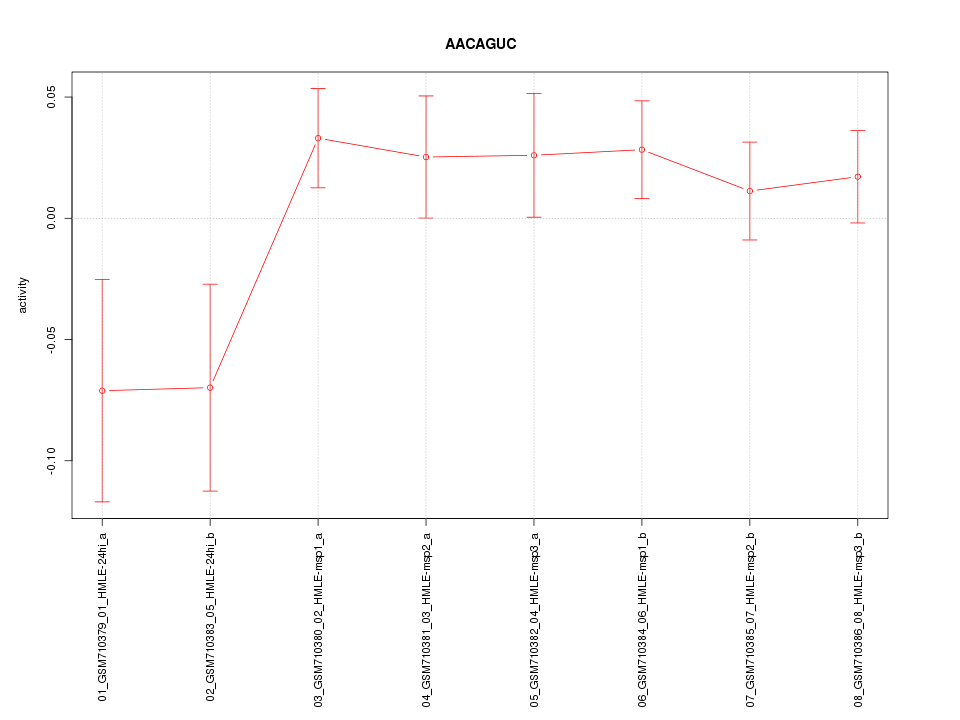
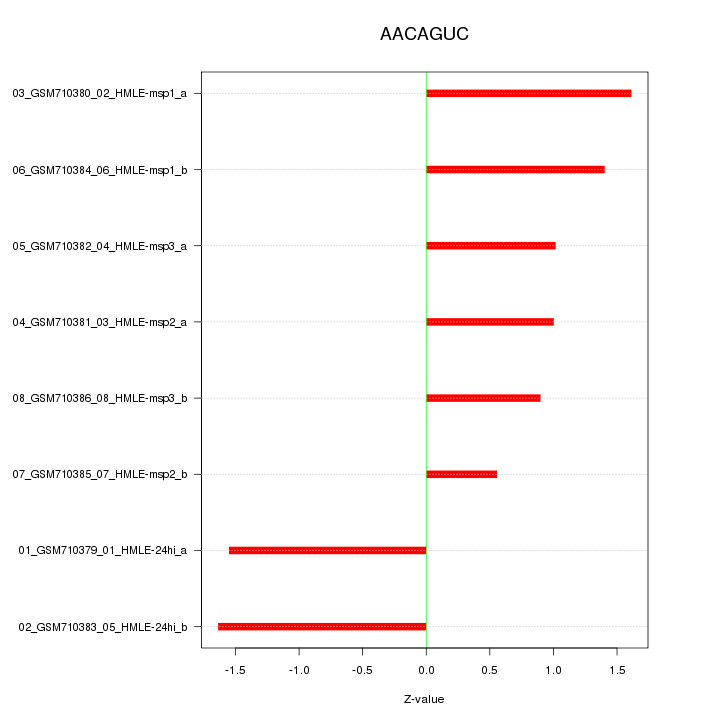
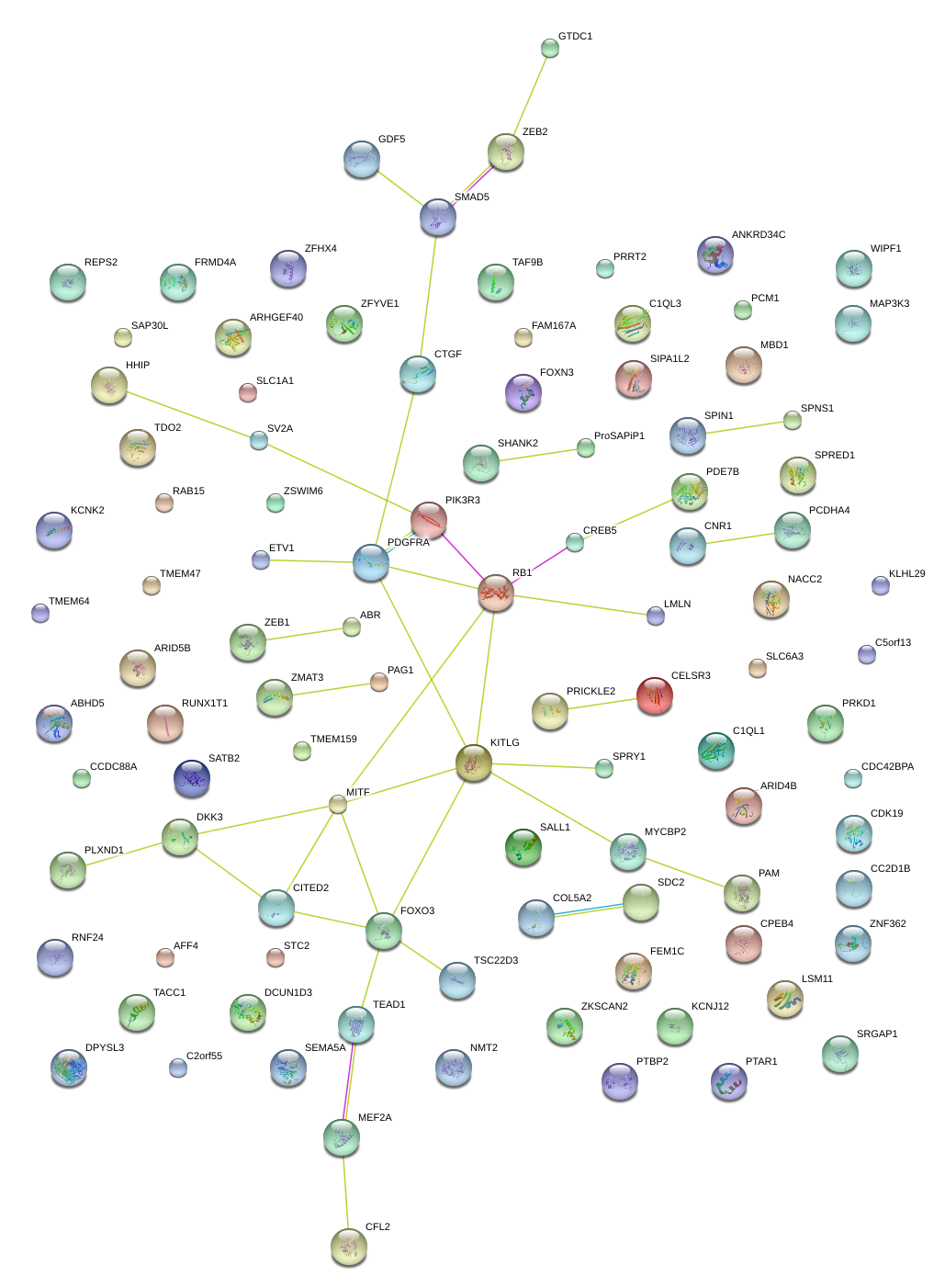
|
chr4_+_145567147
|
2.426
|
NM_022475
|
HHIP
|
hedgehog interacting protein
|
|
chr5_-_111093251
|
2.259
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111312522
|
2.242
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093030
|
2.239
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111092918
|
2.222
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093792
|
2.194
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr20_-_34025955
|
1.474
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr2_-_145277879
|
1.417
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr5_-_111091947
|
1.301
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_-_88875766
|
1.155
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr14_-_89883306
|
1.120
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr4_+_124317884
|
1.117
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr4_+_124320664
|
1.079
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr11_-_70935807
|
1.003
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr11_-_70507911
|
1.002
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr2_-_190044330
|
0.978
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr14_-_90085325
|
0.949
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr5_-_146833150
|
0.919
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr1_+_215178884
|
0.913
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr5_-_146889618
|
0.903
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr1_+_215256559
|
0.899
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr14_-_30396811
|
0.894
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr9_-_138987096
|
0.867
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr6_+_108882068
|
0.702
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr5_+_153825497
|
0.691
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr6_-_132272311
|
0.612
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr6_+_108881010
|
0.602
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr16_+_29823408
|
0.573
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr3_+_69812961
|
0.558
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_69812620
|
0.556
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_69915374
|
0.553
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr8_-_93029864
|
0.550
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_+_69788562
|
0.544
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr8_-_93115453
|
0.535
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_-_48700317
|
0.534
|
NM_001407
|
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr5_-_114880536
|
0.532
|
NM_020177
|
FEM1C
|
fem-1 homolog c (C. elegans)
|
|
chr6_-_88854989
|
0.529
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr8_+_77593506
|
0.526
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chrX_-_107019001
|
0.507
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr6_-_139695349
|
0.501
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr5_-_9546157
|
0.498
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr6_-_139695784
|
0.496
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr6_-_111136407
|
0.482
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr14_+_21538339
|
0.478
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr6_+_136172802
|
0.470
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr2_-_200335988
|
0.469
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr8_-_93111915
|
0.465
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_200329810
|
0.456
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chrX_-_106960268
|
0.456
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr2_-_200322699
|
0.440
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr8_-_93107442
|
0.435
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107881
|
0.433
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_149889362
|
0.425
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr4_+_55095263
|
0.397
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr3_+_69985750
|
0.393
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_-_178789597
|
0.383
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr2_-_145090038
|
0.383
|
NM_001164629
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr17_-_43045612
|
0.381
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chrX_-_106959597
|
0.367
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr3_+_43732358
|
0.365
|
NM_016006
|
ABHD5
|
abhydrolase domain containing 5
|
|
chr1_+_97187174
|
0.359
|
NM_021190
|
PTBP2
|
polypyrimidine tract binding protein 2
|
|
chrX_+_16964730
|
0.358
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr5_-_172756505
|
0.349
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr2_-_144994905
|
0.345
|
NM_024659
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr2_-_145052059
|
0.344
|
NM_001006636
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr3_-_129325175
|
0.334
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr14_-_73453651
|
0.333
|
NM_178441
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr20_-_3149070
|
0.332
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr8_+_97505876
|
0.330
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr17_+_21279667
|
0.328
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr15_+_79575145
|
0.322
|
NM_001146341
|
ANKRD34C
|
ankyrin repeat domain 34C
|
|
chr14_-_73493744
|
0.313
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr2_-_175499275
|
0.310
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr8_-_93075190
|
0.307
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_175547471
|
0.303
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr20_-_3996035
|
0.302
|
NM_007219
NM_001134337
NM_001134338
|
RNF24
|
ring finger protein 24
|
|
chr3_-_64211111
|
0.300
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr16_-_51184507
|
0.297
|
NM_001127892
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr15_+_38544966
|
0.294
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr8_-_11324184
|
0.289
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr2_-_99552677
|
0.287
|
NM_207362
|
C2orf55
|
chromosome 2 open reading frame 55
|
|
chr5_+_157170702
|
0.283
|
NM_173491
|
LSM11
|
LSM11, U7 small nuclear RNA associated
|
|
chr7_-_14025826
|
0.281
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr3_+_197687070
|
0.277
|
NM_001136049
NM_033029
|
LMLN
|
leishmanolysin-like (metallopeptidase M8 family)
|
|
chr16_-_20911560
|
0.268
|
NM_173475
|
DCUN1D3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr7_-_14031049
|
0.265
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr7_-_14029641
|
0.261
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr7_+_28475233
|
0.257
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr17_+_61699786
|
0.257
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr7_-_14030864
|
0.256
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr16_-_51185150
|
0.249
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr8_-_91657898
|
0.248
|
NM_001008495
NM_001146273
|
TMEM64
|
transmembrane protein 64
|
|
chr10_-_15210610
|
0.243
|
NM_004808
|
NMT2
|
N-myristoyltransferase 2
|
|
chr7_+_28725597
|
0.243
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr11_+_12695851
|
0.241
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr1_+_33722158
|
0.232
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr1_-_232651227
|
0.231
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr5_+_60628085
|
0.231
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr5_-_1445510
|
0.230
|
NM_001044
|
SLC6A3
|
solute carrier family 6 (neurotransmitter transporter, dopamine), member 3
|
|
chr1_-_227505796
|
0.227
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chr2_-_55646946
|
0.224
|
NM_001135597
NM_018084
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr5_+_102201526
|
0.219
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr2_+_23608297
|
0.218
|
NM_052920
|
KLHL29
|
kelch-like 29 (Drosophila)
|
|
chr8_+_38585703
|
0.212
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chrX_-_77395108
|
0.211
|
NM_015975
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa
|
|
chr5_+_173315313
|
0.209
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr9_+_91003238
|
0.205
|
NM_006717
|
SPIN1
|
spindlin 1
|
|
chr12_+_64238511
|
0.202
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr10_-_14372807
|
0.198
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr1_-_52831817
|
0.198
|
NM_032449
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr8_+_38644721
|
0.195
|
NM_001122824
NM_006283
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr10_+_63661012
|
0.193
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr5_+_135468522
|
0.193
|
NM_001001419
NM_001001420
NM_005903
|
SMAD5
|
SMAD family member 5
|
|
chr8_+_17780351
|
0.189
|
NM_006197
|
PCM1
|
pericentriolar material 1
|
|
chrX_-_34675366
|
0.189
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr11_-_12030128
|
0.188
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr9_-_72374855
|
0.187
|
NM_001099666
|
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1
|
|
chr1_-_46598250
|
0.186
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr14_-_65438794
|
0.185
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chr5_-_132299263
|
0.181
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr8_-_82024277
|
0.178
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr16_-_25268841
|
0.178
|
NM_001012981
|
ZKSCAN2
|
zinc finger with KRAB and SCAN domains 2
|
|
chr7_+_28338939
|
0.175
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr16_+_21169911
|
0.173
|
NM_020422
|
TMEM159
|
transmembrane protein 159
|
|
chr13_+_48877775
|
0.173
|
NM_000321
|
RB1
|
retinoblastoma 1
|
|
chr9_+_4490193
|
0.171
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr14_-_35183722
|
0.170
|
NM_001243645
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr17_-_1090562
|
0.167
|
NM_001159746
|
ABR
|
active BCR-related gene
|
|
chr12_-_88974094
|
0.166
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr1_-_235490801
|
0.164
|
NM_001206794
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr15_+_100173182
|
0.163
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr1_-_101360405
|
0.162
|
NM_001033025
NM_001439
|
EXTL2
|
exostoses (multiple)-like 2
|
|
chr12_-_76953561
|
0.162
|
NM_001003712
NM_020841
|
OSBPL8
|
oxysterol binding protein-like 8
|
|
chr14_-_35182944
|
0.162
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr3_+_180630054
|
0.161
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr17_-_1083008
|
0.160
|
NM_021962
|
ABR
|
active BCR-related gene
|
|
chr1_+_23037329
|
0.160
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr12_+_51632507
|
0.159
|
NM_001136264
NM_001136266
NM_001136267
NM_001136268
NM_001136269
NM_014764
|
DAZAP2
|
DAZ associated protein 2
|
|
chr6_+_11538486
|
0.159
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr17_-_37557851
|
0.159
|
NM_001184906
NM_032875
|
FBXL20
|
F-box and leucine-rich repeat protein 20
|
|
chr11_-_12030882
|
0.159
|
NM_013253
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr11_-_33744272
|
0.159
|
NM_001127223
NM_001127225
NM_001127226
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr5_-_180688095
|
0.157
|
NM_032765
|
TRIM52
|
tripartite motif containing 52
|
|
chr5_+_122424840
|
0.156
|
NM_001136239
|
PRDM6
|
PR domain containing 6
|
|
chr20_+_56884749
|
0.156
|
NM_020673
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr2_+_12856813
|
0.155
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr11_-_12030622
|
0.155
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr10_+_69644938
|
0.153
|
NM_001142498
|
SIRT1
|
sirtuin 1
|
|
chr17_-_1012257
|
0.153
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chrX_-_103087106
|
0.150
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr15_-_37390372
|
0.149
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr20_+_56964162
|
0.149
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr2_+_95831176
|
0.147
|
NM_001017396
NM_021088
|
ZNF2
|
zinc finger protein 2
|
|
chr12_-_76478737
|
0.147
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr17_-_16118789
|
0.147
|
NM_006311
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr11_-_33744497
|
0.147
|
NM_001127227
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr2_+_48757292
|
0.146
|
NM_001198595
NM_006873
|
STON1
|
stonin 1
|
|
chr9_+_129622935
|
0.146
|
NM_001099270
|
ZBTB34
|
zinc finger and BTB domain containing 34
|
|
chr9_+_72658496
|
0.141
|
NM_153267
|
MAMDC2
|
MAM domain containing 2
|
|
chr5_-_131826426
|
0.139
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr10_+_89623091
|
0.136
|
NM_000314
|
PTEN
|
phosphatase and tensin homolog
|
|
chr14_-_61190458
|
0.135
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chrX_+_109246338
|
0.134
|
NM_032227
|
TMEM164
|
transmembrane protein 164
|
|
chr13_+_100258886
|
0.134
|
NM_206808
|
CLYBL
|
citrate lyase beta like
|
|
chr7_-_111846384
|
0.133
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr10_-_21814610
|
0.131
|
NM_207371
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr9_-_14398981
|
0.130
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr3_+_42642105
|
0.129
|
NM_005385
|
NKTR
|
natural killer-tumor recognition sequence
|
|
chr11_+_12399024
|
0.128
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr7_-_2883649
|
0.128
|
NM_007353
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12
|
|
chr12_+_99038918
|
0.128
|
NM_001160
NM_013229
NM_181861
NM_181868
NM_181869
|
APAF1
|
apoptotic peptidase activating factor 1
|
|
chr9_+_137967088
|
0.127
|
NM_014279
NM_006334
|
OLFM1
|
olfactomedin 1
|
|
chr17_-_73150769
|
0.127
|
NM_001002032
NM_001002033
NM_016185
|
HN1
|
hematological and neurological expressed 1
|
|
chr19_-_51845377
|
0.127
|
NM_001163922
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like
|
|
chr4_+_7045142
|
0.127
|
NM_152293
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr8_-_116681103
|
0.126
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr14_-_54955714
|
0.126
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr7_+_90338711
|
0.125
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr7_+_28452143
|
0.125
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr2_-_198650937
|
0.123
|
NM_197970
|
BOLL
|
bol, boule-like (Drosophila)
|
|
chr11_-_33757872
|
0.123
|
NM_000611
NM_203329
NM_203330
NM_203331
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr7_-_105319608
|
0.120
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr2_-_157189040
|
0.120
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr5_-_64777703
|
0.120
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr1_+_223889253
|
0.120
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr15_-_60919642
|
0.120
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_+_100106132
|
0.117
|
NM_001130926
NM_001130927
NM_001171894
NM_005587
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr21_-_28339405
|
0.116
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr12_+_107168336
|
0.116
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr7_-_100493481
|
0.116
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr12_+_60083125
|
0.115
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr8_+_17013799
|
0.114
|
NM_016353
|
ZDHHC2
|
zinc finger, DHHC-type containing 2
|
|
chr19_+_18794391
|
0.113
|
NM_001098482
NM_015321
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr5_-_38556682
|
0.113
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr8_-_134584012
|
0.110
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr13_-_50510410
|
0.109
|
NM_001127482
NM_020456
|
SPRYD7
|
SPRY domain containing 7
|
|
chr5_-_38595506
|
0.109
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr12_-_101801499
|
0.108
|
NM_001177
|
ARL1
|
ADP-ribosylation factor-like 1
|
|
chr4_-_164253946
|
0.107
|
NM_000909
|
NPY1R
|
neuropeptide Y receptor Y1
|
|
chr10_+_114206708
|
0.107
|
NM_145206
|
VTI1A
|
vesicle transport through interaction with t-SNAREs homolog 1A (yeast)
|