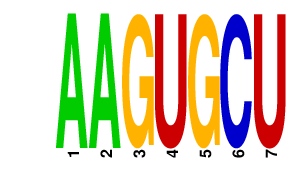
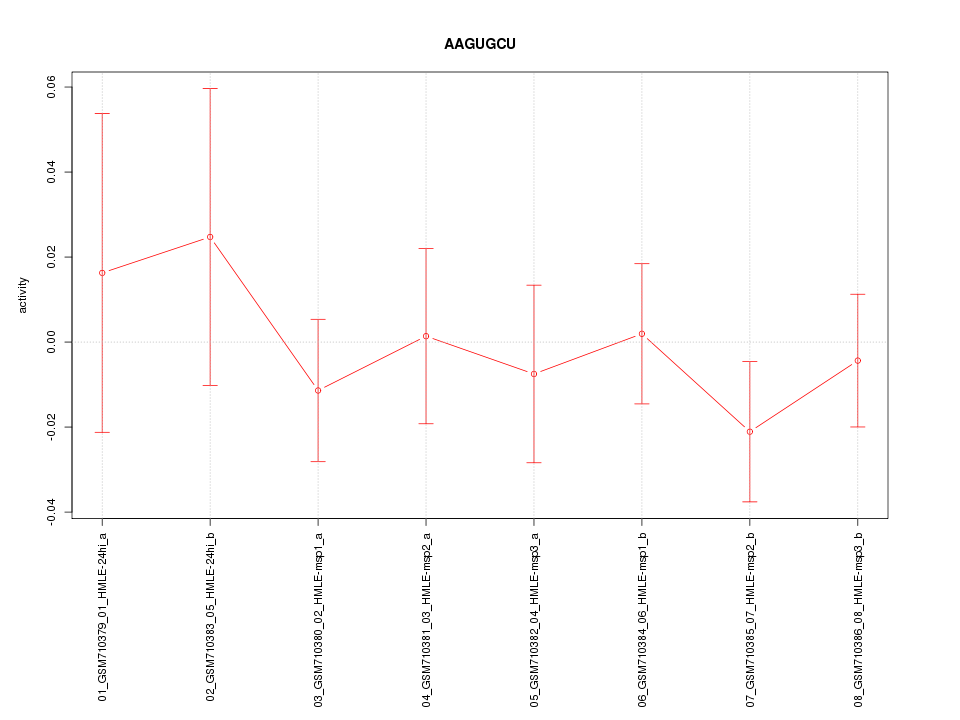
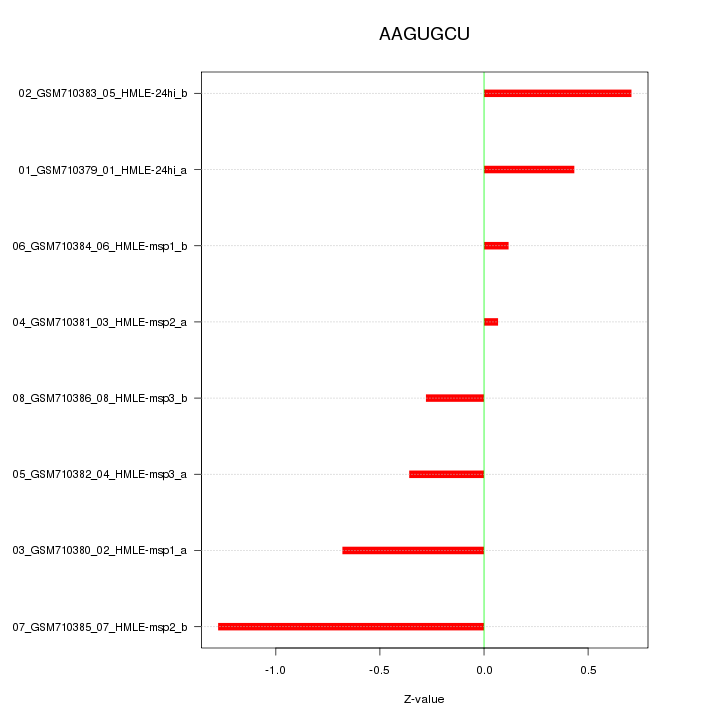
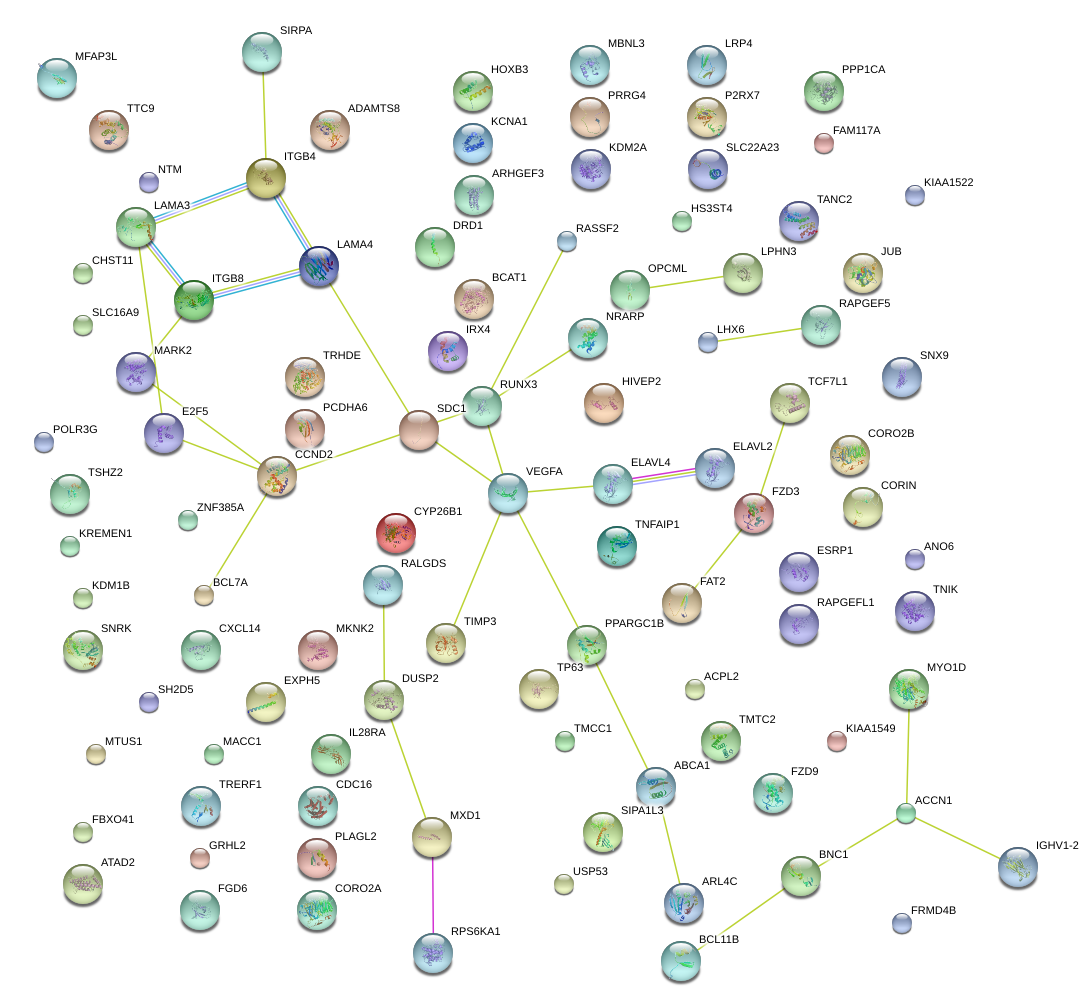
|
chr18_+_21452981
|
1.089
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr8_+_95653340
|
0.911
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr3_-_56809594
|
0.789
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr3_-_56835829
|
0.778
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr3_-_57113292
|
0.724
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr8_+_102504661
|
0.668
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr8_+_28351694
|
0.633
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr12_+_4382882
|
0.620
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr9_-_23826062
|
0.587
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr9_-_124984018
|
0.567
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr9_-_23821842
|
0.544
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr9_-_23821477
|
0.526
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr9_-_124990706
|
0.485
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chr1_-_21059132
|
0.470
|
NM_001103160
NM_001103161
|
SH2D5
|
SH2 domain containing 5
|
|
chr9_-_124990949
|
0.455
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr12_+_83080901
|
0.447
|
NM_152588
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2
|
|
chr2_-_235405692
|
0.446
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr17_-_31203889
|
0.420
|
NM_015194
|
MYO1D
|
myosin ID
|
|
chr12_-_54778456
|
0.416
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr7_-_22396511
|
0.406
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr17_+_38334241
|
0.405
|
NM_016339
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1
|
|
chr12_-_54785018
|
0.398
|
NM_001130967
NM_001130968
|
ZNF385A
|
zinc finger protein 385A
|
|
chr5_-_1882764
|
0.398
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr17_+_73717515
|
0.395
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr3_+_189349215
|
0.372
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr11_-_108464344
|
0.370
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr8_-_17579729
|
0.369
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr9_-_124989755
|
0.366
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr14_-_99737821
|
0.360
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr3_+_189507448
|
0.351
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr8_-_17555158
|
0.348
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr8_-_17580024
|
0.345
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr7_+_20370724
|
0.343
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr7_-_20256942
|
0.343
|
NM_182762
|
MACC1
|
metastasis associated in colon cancer 1
|
|
chr2_-_96811138
|
0.338
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr2_-_20425166
|
0.331
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr8_-_17658385
|
0.327
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr5_+_149109814
|
0.323
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr4_+_120133764
|
0.323
|
NM_019050
|
USP53
|
ubiquitin specific peptidase 53
|
|
chr2_+_85360373
|
0.312
|
NM_031283
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box)
|
|
chr5_+_149151502
|
0.310
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr3_+_140950671
|
0.308
|
NM_001037172
NM_152282
|
ACPL2
|
acid phosphatase-like 2
|
|
chr2_-_72374919
|
0.306
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr2_-_20424627
|
0.296
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr20_+_1875382
|
0.289
|
NM_080792
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr5_+_89770680
|
0.288
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr20_+_1875825
|
0.280
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr6_+_43737937
|
0.278
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr1_+_33231227
|
0.274
|
NM_001198972
NM_001198973
|
KIAA1522
|
KIAA1522
|
|
chr5_-_134914968
|
0.272
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr3_-_69435429
|
0.266
|
NM_015123
|
FRMD4B
|
FERM domain containing 4B
|
|
chr17_+_73720775
|
0.264
|
NM_001005619
|
ITGB4
|
integrin, beta 4
|
|
chr17_-_47841517
|
0.258
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr4_-_170947428
|
0.258
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr20_+_1874779
|
0.257
|
NM_001040022
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr15_-_83953465
|
0.253
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr12_+_104850641
|
0.249
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr12_+_5019048
|
0.248
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr22_+_33196801
|
0.237
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr6_-_42419782
|
0.237
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr6_+_18155535
|
0.231
|
NM_153042
|
KDM1B
|
lysine (K)-specific demethylase 1B
|
|
chr1_-_25256767
|
0.228
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr1_+_33207343
|
0.227
|
NM_020888
|
KIAA1522
|
KIAA1522
|
|
chr12_-_95611023
|
0.224
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr4_-_170924911
|
0.220
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr7_-_138665853
|
0.215
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr12_+_72666462
|
0.212
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr17_-_46651809
|
0.210
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr12_+_122459791
|
0.209
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr18_+_21269528
|
0.204
|
NM_001127717
NM_198129
|
LAMA3
|
laminin, alpha 3
|
|
chr6_-_3456734
|
0.200
|
NM_015482
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr20_+_51588945
|
0.195
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr1_+_26856240
|
0.192
|
NM_002953
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr8_+_86099909
|
0.189
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr20_+_51801821
|
0.185
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr9_-_100935173
|
0.184
|
NM_003389
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr1_+_26872342
|
0.183
|
NM_001006665
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr8_+_86089618
|
0.183
|
NM_001083588
NM_001951
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr3_-_171177851
|
0.177
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr14_+_71108373
|
0.174
|
NM_015351
|
TTC9
|
tetratricopeptide repeat domain 9
|
|
chr6_-_143266283
|
0.171
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr15_+_68871285
|
0.164
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr3_+_43328001
|
0.162
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr19_-_2051222
|
0.161
|
NM_017572
NM_199054
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr9_-_107690526
|
0.161
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr9_-_100954883
|
0.156
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr5_-_174871162
|
0.156
|
NM_000794
|
DRD1
|
dopamine receptor D1
|
|
chr2_+_70142152
|
0.155
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr2_-_73496595
|
0.155
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr6_+_158244281
|
0.154
|
NM_016224
|
SNX9
|
sorting nexin 9
|
|
chr6_-_3445198
|
0.153
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr12_+_45686465
|
0.152
|
NM_001142678
|
ANO6
|
anoctamin 6
|
|
chr16_+_25703346
|
0.151
|
NM_006040
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4
|
|
chr9_-_140196702
|
0.150
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr20_-_30795346
|
0.150
|
NM_002657
|
PLAGL2
|
pleiomorphic adenoma gene-like 2
|
|
chr15_+_68908878
|
0.149
|
NM_001190456
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr8_-_124408688
|
0.147
|
NM_014109
|
ATAD2
|
ATPase family, AAA domain containing 2
|
|
chr17_+_26662547
|
0.146
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr9_-_135996287
|
0.144
|
NM_006266
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr10_-_61469322
|
0.142
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr22_+_29468887
|
0.140
|
NM_001039570
NM_032045
|
KREMEN1
|
kringle containing transmembrane protein 1
|
|
chr11_+_32851469
|
0.138
|
NM_024081
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane)
|
|
chr9_-_136024522
|
0.135
|
NM_001042368
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr19_+_38397841
|
0.134
|
NM_015073
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3
|
|
chr11_+_63606346
|
0.134
|
NM_001039469
NM_001163296
NM_001163297
NM_004954
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr11_+_66886735
|
0.134
|
NM_012308
|
KDM2A
|
lysine (K)-specific demethylase 2A
|
|
chr3_-_129612371
|
0.133
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr14_-_23451704
|
0.133
|
NM_032876
|
AJUBA
|
ajuba LIM protein
|
|
chr11_-_46939908
|
0.133
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr15_+_68924326
|
0.133
|
NM_001190457
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr1_-_24513737
|
0.131
|
NM_170743
NM_173064
NM_173065
|
IL28RA
|
interleukin 28 receptor, alpha (interferon, lambda receptor)
|
|
chr20_-_4795768
|
0.131
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr17_+_61086897
|
0.131
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chrX_-_131623994
|
0.130
|
NM_001170704
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr5_+_140247767
|
0.128
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr12_-_25055966
|
0.127
|
NM_001178093
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr1_-_25291474
|
0.127
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr11_-_133402227
|
0.124
|
NM_001012393
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr5_+_140220811
|
0.123
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr5_+_140254930
|
0.123
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr12_-_25055228
|
0.122
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr2_+_29338207
|
0.122
|
NM_024692
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4
|
|
chr12_-_25102281
|
0.121
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr5_+_140213968
|
0.120
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr3_+_126707380
|
0.120
|
NM_032242
|
PLXNA1
|
plexin A1
|
|
chr1_-_114354906
|
0.120
|
NM_018364
|
RSBN1
|
round spermatid basic protein 1
|
|
chr10_+_98740987
|
0.118
|
NM_015652
|
C10orf12
|
chromosome 10 open reading frame 12
|
|
chr21_-_43346780
|
0.117
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr5_+_140186658
|
0.117
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr1_-_116383323
|
0.117
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr12_-_48298765
|
0.115
|
NM_000376
NM_001017535
NM_001017536
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor
|
|
chr2_+_103236147
|
0.115
|
NM_003048
|
SLC9A2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2
|
|
chrX_-_131573638
|
0.114
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr10_+_112631552
|
0.113
|
NM_001199492
NM_014456
NM_145341
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr22_-_42611310
|
0.113
|
NM_005650
NM_181492
|
TCF20
|
transcription factor 20 (AR1)
|
|
chr17_+_54911452
|
0.111
|
NM_003647
|
DGKE
|
diacylglycerol kinase, epsilon 64kDa
|
|
chr4_+_81118639
|
0.110
|
NM_001099403
|
PRDM8
|
PR domain containing 8
|
|
chr3_-_129599220
|
0.110
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr10_-_90342947
|
0.109
|
NM_001031709
NM_018363
|
RNLS
|
renalase, FAD-dependent amine oxidase
|
|
chr17_+_4736612
|
0.109
|
NM_001024937
NM_015716
NM_153827
NM_170663
|
MINK1
|
misshapen-like kinase 1
|
|
chr5_+_56110899
|
0.109
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr3_-_38691118
|
0.107
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr16_-_4323000
|
0.106
|
NM_003223
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chr1_+_167189948
|
0.104
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr4_+_38665587
|
0.103
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr6_-_108395939
|
0.103
|
NM_014028
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr20_-_31071187
|
0.103
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr1_-_116383692
|
0.102
|
NM_005599
|
NHLH2
|
nescient helix loop helix 2
|
|
chr19_+_11649578
|
0.102
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr20_-_4804067
|
0.101
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr5_+_140345746
|
0.101
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr22_+_29279573
|
0.101
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr14_-_45431121
|
0.101
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr3_-_123339423
|
0.100
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr11_+_22359666
|
0.099
|
NM_020346
|
SLC17A6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6
|
|
chr6_-_16761600
|
0.099
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chrX_-_15353675
|
0.098
|
NM_002641
NM_020473
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr4_+_86699846
|
0.097
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chrX_+_24483343
|
0.097
|
NM_001142386
NM_005391
|
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3
|
|
chr10_+_23728197
|
0.096
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chrX_-_131547536
|
0.096
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr12_-_16760935
|
0.096
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr1_+_32645322
|
0.095
|
NM_175852
|
TXLNA
|
taxilin alpha
|
|
chr3_-_10547267
|
0.095
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr10_-_86001197
|
0.094
|
NM_015613
|
LRIT1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1
|
|
chr2_-_60780404
|
0.093
|
NM_018014
NM_022893
NM_138559
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr12_+_19592311
|
0.093
|
NM_001114176
NM_153207
|
AEBP2
|
AE binding protein 2
|
|
chr14_-_23446431
|
0.093
|
NM_198086
|
AJUBA
|
ajuba LIM protein
|
|
chr3_-_121264780
|
0.092
|
NM_199420
|
POLQ
|
polymerase (DNA directed), theta
|
|
chr3_-_123603144
|
0.092
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr5_+_140207562
|
0.091
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr1_-_45956795
|
0.091
|
NM_007170
|
TESK2
|
testis-specific kinase 2
|
|
chrX_-_20284708
|
0.090
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr18_+_42260774
|
0.090
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr21_-_43373938
|
0.090
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr4_-_185395616
|
0.090
|
NM_002199
|
IRF2
|
interferon regulatory factor 2
|
|
chr17_+_18086813
|
0.089
|
NM_017758
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr11_-_132812870
|
0.089
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr14_+_33408458
|
0.088
|
NM_001164749
NM_001165893
NM_022123
NM_173159
|
NPAS3
|
neuronal PAS domain protein 3
|
|
chr4_+_99916787
|
0.088
|
NM_015143
|
METAP1
|
methionyl aminopeptidase 1
|
|
chr6_-_24383519
|
0.087
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr12_-_51591949
|
0.086
|
NM_002702
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr1_-_156051788
|
0.085
|
NM_001093725
|
MEX3A
|
mex-3 homolog A (C. elegans)
|
|
chr18_+_55019720
|
0.085
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr11_-_102323353
|
0.085
|
NM_052932
|
TMEM123
|
transmembrane protein 123
|
|
chr7_-_75368276
|
0.085
|
NM_001243198
NM_005338
|
HIP1
|
huntingtin interacting protein 1
|
|
chrX_+_105412297
|
0.084
|
NM_001171020
NM_152423
|
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1
|
|
chr12_+_45609767
|
0.084
|
NM_001025356
NM_001142679
NM_001204803
|
ANO6
|
anoctamin 6
|
|
chrX_+_48555124
|
0.083
|
NM_003173
|
SUV39H1
|
suppressor of variegation 3-9 homolog 1 (Drosophila)
|
|
chr13_-_20735182
|
0.083
|
NM_021954
|
GJA3
|
gap junction protein, alpha 3, 46kDa
|
|
chr11_-_65381641
|
0.083
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr13_-_26795821
|
0.083
|
NM_183044
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr1_+_245318245
|
0.082
|
NM_018012
|
KIF26B
|
kinesin family member 26B
|
|
chr3_+_14860468
|
0.080
|
NM_152536
|
FGD5
|
FYVE, RhoGEF and PH domain containing 5
|
|
chr4_+_81106423
|
0.080
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr1_-_44482996
|
0.080
|
NM_006934
NM_201649
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr13_-_26795927
|
0.080
|
NM_183043
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr5_+_140180782
|
0.080
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr1_+_2985730
|
0.080
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr17_+_36861857
|
0.079
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|