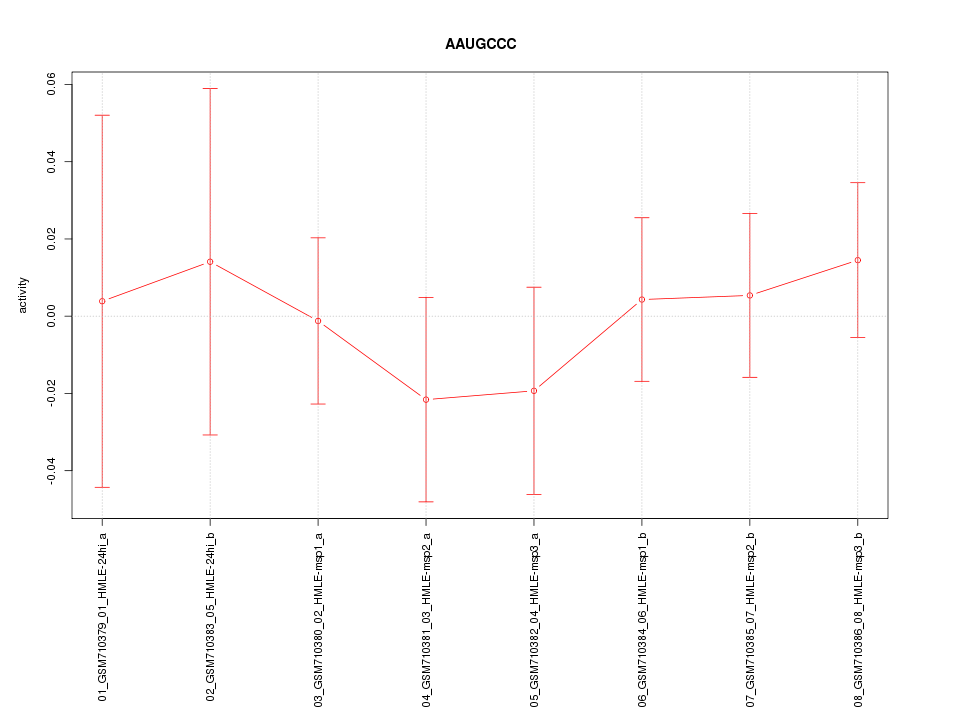
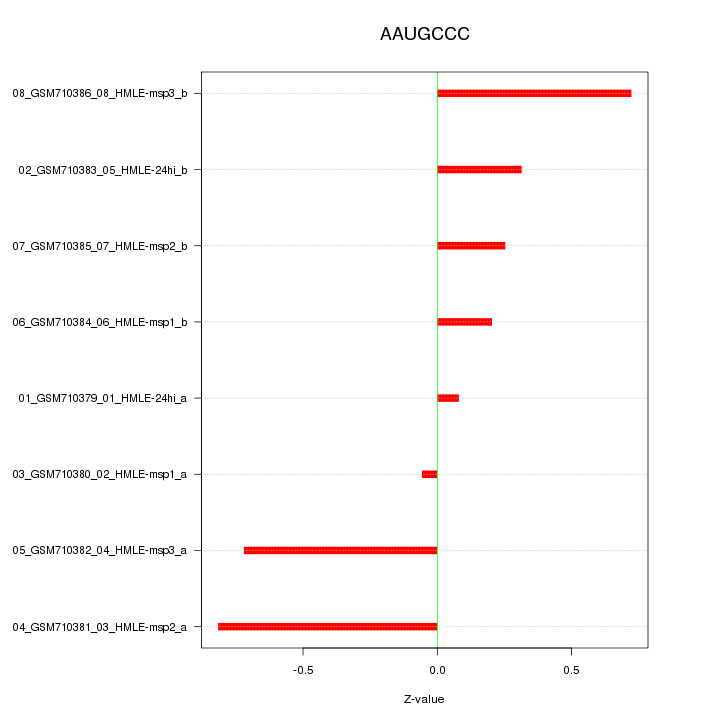
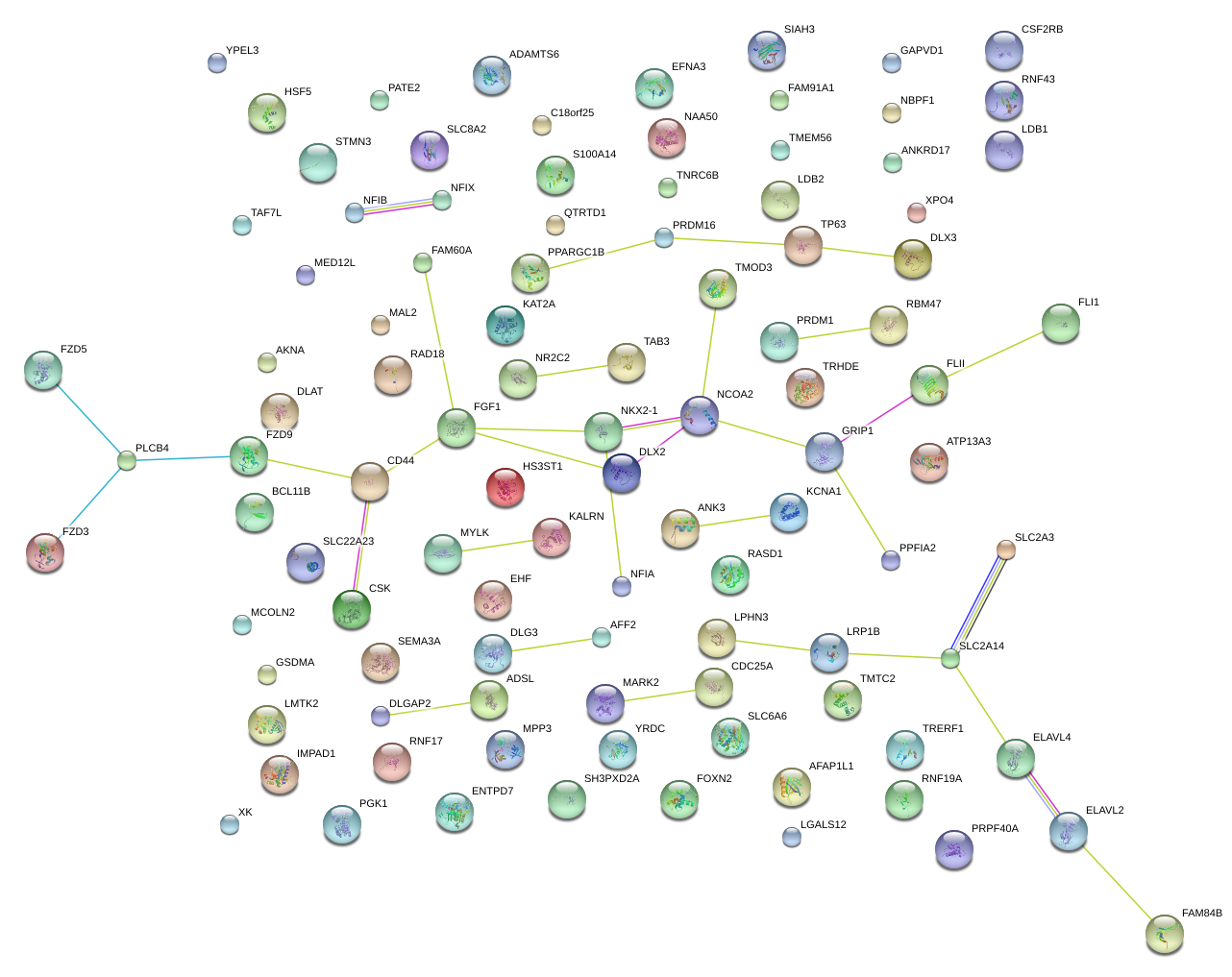
|
chr5_+_149109814
|
0.222
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr5_+_149151502
|
0.220
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr1_-_153588789
|
0.206
|
NM_020672
|
S100A14
|
S100 calcium binding protein A14
|
|
chr11_+_34654010
|
0.162
|
NM_001206616
|
EHF
|
ets homologous factor
|
|
chr12_+_72666462
|
0.159
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr8_+_28351694
|
0.158
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr11_+_34642587
|
0.150
|
NM_001206615
NM_012153
|
EHF
|
ets homologous factor
|
|
chr3_-_123339423
|
0.137
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr13_-_21476895
|
0.126
|
NM_022459
|
XPO4
|
exportin 4
|
|
chr10_-_105614952
|
0.120
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr3_-_123603144
|
0.115
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr8_-_127570710
|
0.097
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr9_-_23826062
|
0.089
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr7_-_83824216
|
0.087
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr3_-_48229800
|
0.082
|
NM_001789
NM_201567
|
CDC25A
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr12_+_5019048
|
0.080
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chrX_+_37545008
|
0.079
|
NM_021083
|
XK
|
X-linked Kx blood group (McLeod syndrome)
|
|
chr9_-_23821477
|
0.079
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr1_-_85462622
|
0.078
|
NM_153259
|
MCOLN2
|
mucolipin 2
|
|
chr3_+_189507448
|
0.077
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr14_-_99737821
|
0.077
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr9_-_23821842
|
0.076
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr11_-_125648704
|
0.075
|
NM_212555
|
PATE2
|
prostate and testis expressed 2
|
|
chr2_-_208634051
|
0.074
|
NM_003468
|
FZD5
|
frizzled family receptor 5
|
|
chr12_-_67072881
|
0.074
|
NM_001178074
NM_021150
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chr11_+_128563772
|
0.073
|
NM_001167681
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr3_+_189349215
|
0.073
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr8_+_120220554
|
0.072
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr8_+_124780742
|
0.069
|
NM_144963
|
FAM91A1
|
family with sequence similarity 91, member A1
|
|
chr20_+_9198036
|
0.066
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr7_+_97736118
|
0.066
|
NM_014916
|
LMTK2
|
lemur tyrosine kinase 2
|
|
chr1_-_38273852
|
0.066
|
NM_024640
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr20_+_9288446
|
0.065
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr5_-_64777703
|
0.064
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr20_+_9049660
|
0.064
|
NM_001172646
|
PLCB4
|
phospholipase C, beta 4
|
|
chr1_+_95558072
|
0.063
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr4_-_11430502
|
0.063
|
NM_005114
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1
|
|
chr8_+_1449531
|
0.063
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr9_-_117156684
|
0.062
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr2_-_153573858
|
0.062
|
NM_017892
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae)
|
|
chr11_+_63606346
|
0.062
|
NM_001039469
NM_001163296
NM_001163297
NM_004954
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr3_+_124303505
|
0.062
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr11_+_63655986
|
0.061
|
NM_017490
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr10_-_103874661
|
0.059
|
NM_003893
|
LDB1
|
LIM domain binding 1
|
|
chr1_+_95582827
|
0.057
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr3_-_194188967
|
0.056
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr12_-_31479120
|
0.055
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr17_-_56565700
|
0.054
|
NM_001080439
|
HSF5
|
heat shock transcription factor family member 5
|
|
chr15_+_75074417
|
0.053
|
NM_001127190
NM_004383
|
CSK
|
c-src tyrosine kinase
|
|
chr2_-_142889269
|
0.053
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr3_+_14989076
|
0.052
|
NM_003298
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2
|
|
chr17_-_48072566
|
0.051
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr12_-_8088629
|
0.051
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr17_+_38119225
|
0.049
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr18_+_43753986
|
0.049
|
NM_001008239
NM_145055
|
C18orf25
|
chromosome 18 open reading frame 25
|
|
chr10_+_101419169
|
0.047
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr5_-_142065586
|
0.047
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr3_+_113775580
|
0.047
|
NM_024638
|
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1
|
|
chr9_+_128024109
|
0.047
|
NM_015635
|
GAPVD1
|
GTPase activating protein and VPS9 domains 1
|
|
chr3_-_9005070
|
0.046
|
NM_020165
|
RAD18
|
RAD18 homolog (S. cerevisiae)
|
|
chr5_-_142065991
|
0.046
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr10_-_103879933
|
0.046
|
NM_001113407
|
LDB1
|
LIM domain binding 1
|
|
chr17_-_56494930
|
0.046
|
NM_017763
|
RNF43
|
ring finger protein 43
|
|
chr13_-_46425756
|
0.044
|
NM_198849
|
SIAH3
|
seven in absentia homolog 3 (Drosophila)
|
|
chr12_+_83080901
|
0.044
|
NM_152588
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2
|
|
chrX_+_77359665
|
0.044
|
NM_000291
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr3_-_113465101
|
0.043
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr5_-_142000899
|
0.043
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr8_-_57906423
|
0.042
|
NM_017813
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr3_+_150804675
|
0.042
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr11_+_35160385
|
0.042
|
NM_000610
NM_001001389
NM_001001390
NM_001001391
NM_001001392
NM_001202555
NM_001202556
NM_001202557
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr5_-_142077508
|
0.041
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chrX_+_69664704
|
0.041
|
NM_021120
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr4_-_40517913
|
0.041
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr10_-_61900659
|
0.040
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr6_-_42419782
|
0.040
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr5_-_141993691
|
0.040
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr10_-_62149487
|
0.039
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr4_-_74124456
|
0.039
|
NM_032217
NM_198889
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr17_-_40273304
|
0.039
|
NM_021078
|
KAT2A
|
K(lysine) acetyltransferase 2A
|
|
chr6_-_3456734
|
0.039
|
NM_015482
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chrX_-_30907408
|
0.039
|
NM_152787
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3
|
|
chr10_-_62332666
|
0.039
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr5_+_148651395
|
0.038
|
NM_001146337
NM_152406
|
AFAP1L1
|
actin filament associated protein 1-like 1
|
|
chr22_+_37309618
|
0.038
|
NM_000395
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr1_+_155051263
|
0.037
|
NM_004952
|
EFNA3
|
ephrin-A3
|
|
chr1_+_2985730
|
0.036
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr14_-_36988902
|
0.035
|
NM_003317
|
NKX2-1
|
NK2 homeobox 1
|
|
chrX_+_147582133
|
0.035
|
NM_001169122
NM_001169123
NM_001169124
NM_001169125
NM_002025
|
AFF2
|
AF4/FMR2 family, member 2
|
|
chr19_-_47975244
|
0.034
|
NM_015063
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr11_+_111895537
|
0.034
|
NM_001931
|
DLAT
|
dihydrolipoamide S-acetyltransferase
|
|
chr14_-_36989333
|
0.034
|
NM_001079668
|
NKX2-1
|
NK2 homeobox 1
|
|
chr15_+_52121807
|
0.034
|
NM_014547
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr3_+_14444083
|
0.034
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr6_+_106534188
|
0.033
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chrX_+_69674914
|
0.033
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chrX_+_69672110
|
0.033
|
NM_020730
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr20_-_62284701
|
0.033
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr6_+_106546736
|
0.032
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr2_+_48541767
|
0.032
|
NM_002158
|
FOXN2
|
forkhead box N2
|
|
chr22_+_40573879
|
0.032
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chrX_+_147800610
|
0.032
|
NM_001170628
|
AFF2
|
AF4/FMR2 family, member 2
|
|
chr17_-_17399494
|
0.032
|
NM_001199989
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr9_-_14314036
|
0.032
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr12_-_81763127
|
0.031
|
NM_001220479
NM_001220480
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr12_-_123594974
|
0.031
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr4_+_38665587
|
0.031
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr17_-_38574042
|
0.031
|
NM_001067
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa
|
|
chr3_-_47823386
|
0.030
|
NM_003074
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr1_-_23857711
|
0.030
|
NM_004091
|
E2F2
|
E2F transcription factor 2
|
|
chr11_+_64073040
|
0.030
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr8_-_20112691
|
0.029
|
NM_021020
|
LZTS1
|
leucine zipper, putative tumor suppressor 1
|
|
chr6_-_3445198
|
0.029
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr5_+_148724976
|
0.029
|
NM_152407
|
GRPEL2
|
GrpE-like 2, mitochondrial (E. coli)
|
|
chr5_-_132073143
|
0.028
|
NM_007054
|
KIF3A
|
kinesin family member 3A
|
|
chr11_-_128392061
|
0.028
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chrX_-_15353675
|
0.028
|
NM_002641
NM_020473
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr6_+_131456825
|
0.027
|
NM_016377
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr12_+_98909331
|
0.027
|
NM_001032283
NM_001032284
NM_003276
|
TMPO
|
thymopoietin
|
|
chr2_-_64881040
|
0.026
|
NM_014755
|
SERTAD2
|
SERTA domain containing 2
|
|
chr12_-_118406027
|
0.026
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr4_-_40631846
|
0.026
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr4_+_91048683
|
0.026
|
NM_001145065
|
FAM190A
|
family with sequence similarity 190, member A
|
|
chr11_-_128457452
|
0.026
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr3_-_15798057
|
0.025
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr12_-_42538432
|
0.025
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr14_-_88737160
|
0.025
|
NM_138318
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr12_-_81991959
|
0.025
|
NM_001220477
NM_001220478
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr5_+_74632992
|
0.025
|
NM_000859
NM_001130996
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase
|
|
chr14_-_88793250
|
0.025
|
NM_021161
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr12_-_82152269
|
0.025
|
NM_001220475
NM_001220476
NM_001220473
NM_001220474
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr5_-_39074494
|
0.024
|
NM_152756
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr10_+_16478941
|
0.024
|
NM_001001484
NM_030664
|
PTER
|
phosphotriesterase related
|
|
chr22_+_21271619
|
0.024
|
NM_005207
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr19_+_50094911
|
0.023
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chr3_+_50229022
|
0.023
|
NM_000172
NM_144499
|
GNAT1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1
|
|
chr10_-_62493282
|
0.023
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_-_156051788
|
0.023
|
NM_001093725
|
MEX3A
|
mex-3 homolog A (C. elegans)
|
|
chr12_+_124118312
|
0.023
|
NM_001516
|
GTF2H3
|
general transcription factor IIH, polypeptide 3, 34kDa
|
|
chr17_-_27621163
|
0.023
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr6_+_80340971
|
0.022
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr5_-_168006413
|
0.022
|
NM_024594
|
PANK3
|
pantothenate kinase 3
|
|
chr22_+_32340435
|
0.022
|
NM_003405
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr12_-_82153092
|
0.022
|
NM_003625
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr10_-_32217762
|
0.022
|
NM_018287
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr8_+_54793409
|
0.021
|
NM_003702
|
RGS20
|
regulator of G-protein signaling 20
|
|
chr14_-_68162419
|
0.021
|
NM_001252650
NM_016026
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis)
|
|
chr12_-_49449106
|
0.021
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr11_+_68816314
|
0.021
|
NM_139075
|
TPCN2
|
two pore segment channel 2
|
|
chr11_-_122932844
|
0.021
|
NM_006597
NM_153201
|
HSPA8
|
heat shock 70kDa protein 8
|
|
chr14_-_88789531
|
0.021
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr17_+_38444134
|
0.021
|
NM_001254
|
CDC6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr17_-_42275425
|
0.021
|
NM_001098833
NM_020218
|
ATXN7L3
|
ataxin 7-like 3
|
|
chr11_-_27528298
|
0.021
|
NM_018362
|
LIN7C
|
lin-7 homolog C (C. elegans)
|
|
chr11_-_16424391
|
0.020
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr22_+_40440664
|
0.020
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr13_+_30002776
|
0.020
|
NM_015233
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr7_+_141251077
|
0.020
|
NM_018238
|
AGK
|
acylglycerol kinase
|
|
chr3_-_125094003
|
0.020
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr6_-_88411984
|
0.019
|
NM_018064
|
AKIRIN2
|
akirin 2
|
|
chr10_-_120514669
|
0.019
|
NM_153810
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr10_-_103815868
|
0.019
|
NM_024541
|
C10orf76
|
chromosome 10 open reading frame 76
|
|
chr1_+_157963062
|
0.019
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr2_-_206950895
|
0.019
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr19_-_42636559
|
0.018
|
NM_001207025
NM_001207026
NM_001247994
NM_002698
|
POU2F2
|
POU class 2 homeobox 2
|
|
chr11_+_69455837
|
0.018
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr1_+_61547388
|
0.018
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr22_+_42229105
|
0.018
|
NM_004599
|
SREBF2
|
sterol regulatory element binding transcription factor 2
|
|
chr1_+_236958539
|
0.018
|
NM_000254
|
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase
|
|
chr3_+_43328001
|
0.017
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr6_+_34857021
|
0.017
|
NM_015245
|
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A
|
|
chr12_-_89918531
|
0.017
|
NM_003774
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4)
|
|
chr11_+_107461807
|
0.017
|
NM_001130037
NM_018712
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chr16_-_67840464
|
0.017
|
NM_020850
|
RANBP10
|
RAN binding protein 10
|
|
chr10_-_735522
|
0.016
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr4_-_149363498
|
0.016
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr3_-_125314367
|
0.016
|
NM_022776
|
OSBPL11
|
oxysterol binding protein-like 11
|
|
chr11_-_16497917
|
0.016
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr12_+_66218141
|
0.015
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr18_+_32397925
|
0.015
|
NM_001198943
|
DTNA
|
dystrobrevin, alpha
|
|
chr16_+_19422056
|
0.015
|
NM_001105248
NM_001105249
|
TMC5
|
transmembrane channel-like 5
|
|
chr2_+_183580960
|
0.015
|
NM_018981
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr9_+_131445933
|
0.015
|
NM_001122821
|
SET
|
SET nuclear oncogene
|
|
chrX_+_73641084
|
0.015
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr5_-_79286793
|
0.015
|
NM_001010891
NM_001167741
|
MTX3
|
metaxin 3
|
|
chr7_+_4721722
|
0.015
|
NM_001037165
|
FOXK1
|
forkhead box K1
|
|
chr13_+_29598746
|
0.014
|
NM_001033602
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr2_+_192542860
|
0.014
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr6_+_131571298
|
0.014
|
NM_004842
NM_138633
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr9_+_116638544
|
0.014
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr10_-_101190201
|
0.014
|
NM_002079
|
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1)
|
|
chr8_-_41909477
|
0.014
|
NM_001099412
NM_001099413
NM_006766
|
KAT6A
|
K(lysine) acetyltransferase 6A
|
|
chr2_-_183731279
|
0.014
|
NM_001463
|
FRZB
|
frizzled-related protein
|
|
chr12_-_70093064
|
0.013
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr12_+_2162388
|
0.013
|
NM_000719
NM_001129827
NM_001129829
NM_001129830
NM_001129831
NM_001129832
NM_001129833
NM_001129834
NM_001129835
NM_001129836
NM_001129837
NM_001129838
NM_001129839
NM_001129840
NM_001129841
NM_001129842
NM_001129843
NM_001129844
NM_001129846
NM_001167623
NM_001167624
NM_001167625
NM_199460
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr8_+_67624652
|
0.013
|
NM_001033578
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr14_+_52734430
|
0.013
|
NM_000953
|
PTGDR
|
prostaglandin D2 receptor (DP)
|
|
chr2_-_136594749
|
0.013
|
NM_002299
|
LCT
|
lactase
|
|
chr18_+_32398225
|
0.013
|
NM_001198942
NM_001198944
NM_032980
NM_032981
|
DTNA
|
dystrobrevin, alpha
|
|
chr8_+_54764367
|
0.012
|
NM_170587
|
RGS20
|
regulator of G-protein signaling 20
|