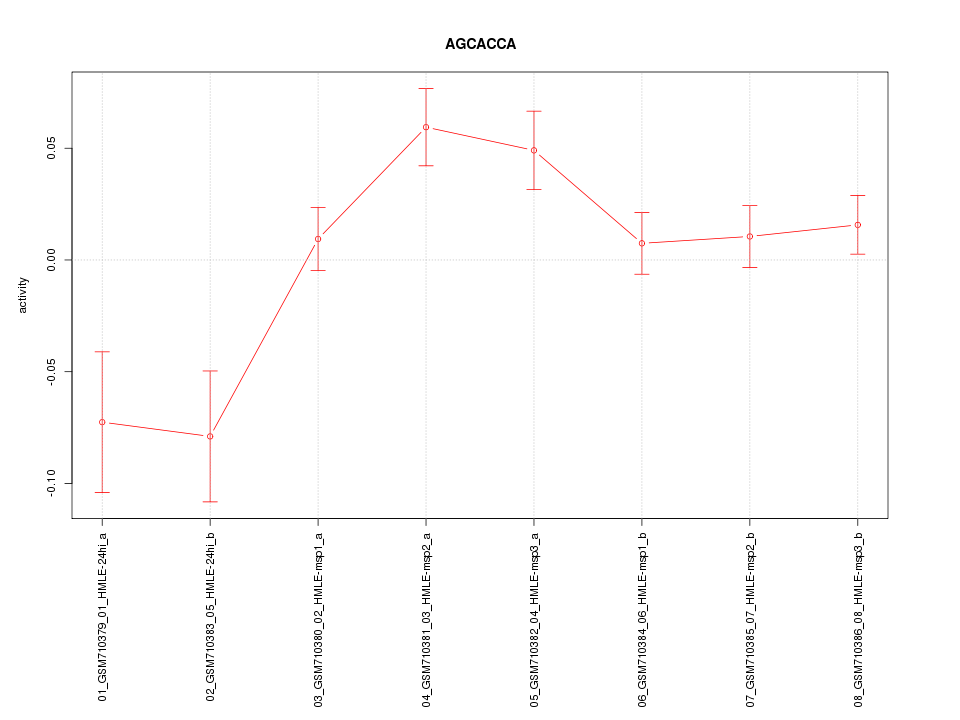
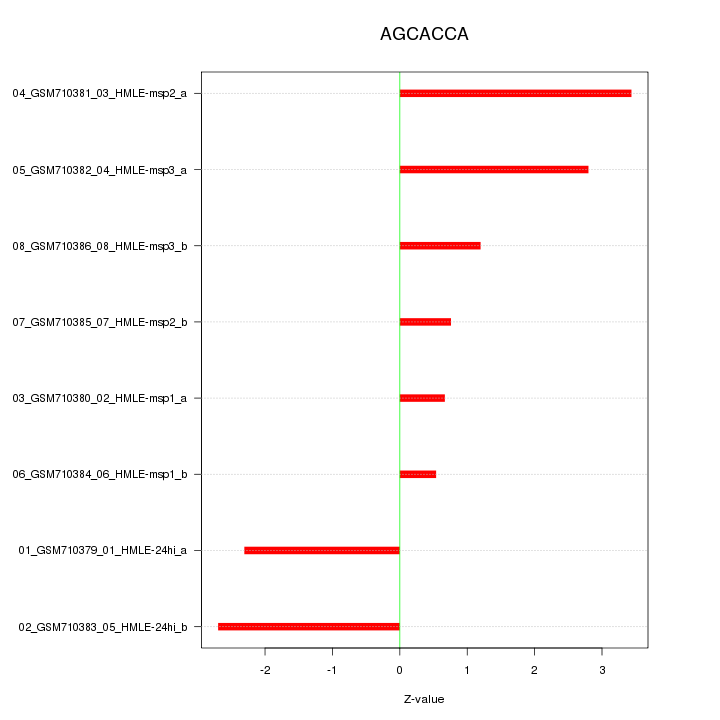
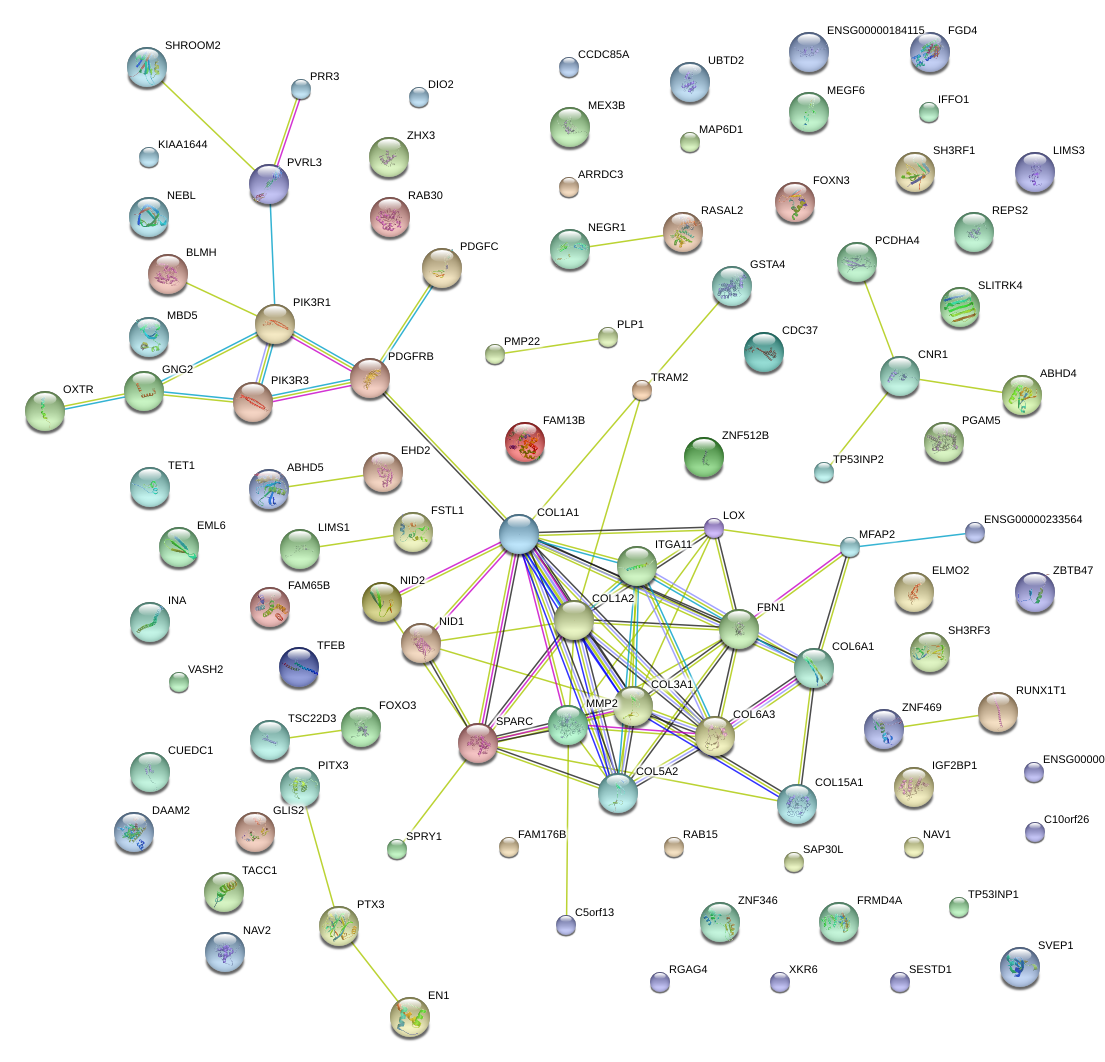
|
chr2_+_189839078
|
6.024
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr7_+_94023872
|
4.834
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr15_-_48937795
|
4.568
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr1_-_236228476
|
4.288
|
NM_002508
|
NID1
|
nidogen 1
|
|
chrX_+_16964730
|
3.490
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr2_+_109745996
|
3.241
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr3_+_157154579
|
3.179
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr5_-_111312522
|
3.128
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093251
|
3.082
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093030
|
3.075
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093792
|
3.049
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111092918
|
3.032
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_-_88875766
|
2.999
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr10_-_21463019
|
2.913
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr5_-_149535420
|
2.731
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr17_-_48278987
|
2.653
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr1_-_3528058
|
2.597
|
NM_001409
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr2_-_190044330
|
2.573
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr16_+_55515468
|
2.539
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr16_+_55512801
|
2.407
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr4_+_124317884
|
2.391
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr4_+_124320664
|
2.328
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr14_-_52535945
|
2.265
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr10_-_21186530
|
2.138
|
NM_006393
|
NEBL
|
nebulette
|
|
chr2_-_238322840
|
2.045
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr5_-_121412805
|
1.937
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr9_+_101705710
|
1.937
|
NM_001855
|
COL15A1
|
collagen, type XV, alpha 1
|
|
chr5_-_121414011
|
1.883
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr22_-_44708730
|
1.827
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr20_+_33292117
|
1.771
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr14_+_52327021
|
1.708
|
NM_001243773
NM_001243774
NM_053064
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr5_-_111091947
|
1.688
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr11_+_19734821
|
1.642
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr1_+_201708940
|
1.641
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chrX_+_9754465
|
1.582
|
NM_001649
|
SHROOM2
|
shroom family member 2
|
|
chr11_+_19372270
|
1.528
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr12_-_6665220
|
1.512
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr1_-_46598250
|
1.459
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr6_-_52441815
|
1.459
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chrX_-_107019001
|
1.456
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr5_+_67511578
|
1.429
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr6_-_88854989
|
1.424
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr14_-_89883306
|
1.424
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr1_+_201617449
|
1.421
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr5_+_67588395
|
1.404
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr5_+_67584251
|
1.384
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_-_72748147
|
1.377
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr5_-_151066492
|
1.357
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr3_-_8811263
|
1.347
|
NM_000916
|
OXTR
|
oxytocin receptor
|
|
chr2_+_56411123
|
1.344
|
NM_001080433
|
CCDC85A
|
coiled-coil domain containing 85A
|
|
chr8_-_95961543
|
1.333
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr5_+_67586466
|
1.328
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chrX_-_142722869
|
1.327
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chrX_-_106960268
|
1.324
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr8_-_93115453
|
1.308
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr6_+_30524485
|
1.303
|
NM_001077497
NM_025263
|
PRR3
|
proline rich 3
|
|
chr16_+_88493878
|
1.295
|
NM_001127464
|
ZNF469
|
zinc finger protein 469
|
|
chr8_-_93029864
|
1.265
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_17308072
|
1.264
|
NM_001135247
NM_017459
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chrX_-_142722318
|
1.261
|
NM_001184750
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr2_-_180129245
|
1.254
|
NM_178123
|
SESTD1
|
SEC14 and spectrin domains 1
|
|
chr6_+_108882068
|
1.230
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr17_+_47074749
|
1.229
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr15_-_82338348
|
1.222
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr14_-_90085325
|
1.222
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chrX_+_103031433
|
1.200
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chrX_+_103031753
|
1.198
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr20_-_39928708
|
1.181
|
NM_015035
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr17_-_28619183
|
1.164
|
NM_000386
|
BLMH
|
bleomycin hydrolase
|
|
chr8_-_93111915
|
1.164
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_17307107
|
1.156
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr3_-_120169493
|
1.145
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr6_-_41702734
|
1.131
|
NM_007162
|
TFEB
|
transcription factor EB
|
|
chr17_-_15165873
|
1.130
|
NM_153321
|
PMP22
|
peripheral myelin protein 22
|
|
chr6_-_41703938
|
1.127
|
NM_001167827
|
TFEB
|
transcription factor EB
|
|
chr8_-_93107881
|
1.123
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_-_142723921
|
1.114
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr17_-_15164043
|
1.113
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chrX_-_106959597
|
1.106
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr17_-_15168590
|
1.103
|
NM_000304
|
PMP22
|
peripheral myelin protein 22
|
|
chr8_-_93107442
|
1.103
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_-_137368778
|
1.070
|
NM_001101800
NM_001101801
NM_016603
|
FAM13B
|
family with sequence similarity 13, member B
|
|
chr6_+_108881010
|
1.057
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr5_+_153825497
|
1.057
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr6_-_52860076
|
1.052
|
NM_001512
|
GSTA4
|
glutathione S-transferase alpha 4
|
|
chr11_-_82782883
|
1.042
|
NM_014488
|
RAB30
|
RAB30, member RAS oncogene family
|
|
chr2_+_148778573
|
1.028
|
NM_018328
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr12_+_32654976
|
1.026
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr4_-_157892545
|
1.012
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr2_+_54952148
|
1.012
|
NM_001039753
|
EML6
|
echinoderm microtubule associated protein like 6
|
|
chr21_+_47401644
|
0.996
|
NM_001848
|
COL6A1
|
collagen, type VI, alpha 1
|
|
chr17_-_55980749
|
0.972
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr10_-_14372807
|
0.971
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr10_+_105036783
|
0.957
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr14_-_65438794
|
0.933
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chrX_-_71351750
|
0.930
|
NM_001024455
|
RGAG4
|
retrotransposon gag domain containing 4
|
|
chr5_-_171710794
|
0.922
|
NM_152277
|
UBTD2
|
ubiquitin domain containing 2
|
|
chr3_+_42695175
|
0.921
|
NM_145166
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr19_+_48216600
|
0.911
|
NM_014601
|
EHD2
|
EH-domain containing 2
|
|
chr14_-_80677839
|
0.911
|
NM_001007023
NM_001242502
NM_001242503
NM_013989
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr1_+_213123853
|
0.883
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr14_+_23067128
|
0.881
|
NM_022060
|
ABHD4
|
abhydrolase domain containing 4
|
|
chr6_+_39760772
|
0.873
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr8_-_93075190
|
0.872
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_+_43732358
|
0.868
|
NM_016006
|
ABHD5
|
abhydrolase domain containing 5
|
|
chr20_-_45035253
|
0.862
|
NM_133171
NM_182764
|
ELMO2
|
engulfment and cell motility 2
|
|
chr16_+_4382224
|
0.853
|
NM_032575
|
GLIS2
|
GLIS family zinc finger 2
|
|
chr8_-_11058847
|
0.852
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr7_-_28998028
|
0.830
|
NM_014817
|
TRIL
|
TLR4 interactor with leucine-rich repeats
|
|
chr10_+_70320044
|
0.829
|
NM_030625
|
TET1
|
tet methylcytosine dioxygenase 1
|
|
chr1_-_36789754
|
0.827
|
NM_018166
|
FAM176B
|
family with sequence similarity 176, member B
|
|
chr2_-_119605758
|
0.825
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr14_-_80697396
|
0.813
|
NM_000793
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr6_+_39760148
|
0.799
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr5_-_90679115
|
0.794
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr15_-_68724434
|
0.793
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr10_+_104503724
|
0.790
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr3_-_183543295
|
0.787
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr8_+_38585703
|
0.782
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr9_-_113341772
|
0.774
|
NM_153366
|
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1
|
|
chr6_-_24911194
|
0.769
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr20_-_62601140
|
0.763
|
NM_020713
|
ZNF512B
|
zinc finger protein 512B
|
|
chr5_+_176449696
|
0.745
|
NM_012279
|
ZNF346
|
zinc finger protein 346
|
|
chr2_+_109237679
|
0.735
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr8_+_38644721
|
0.734
|
NM_001122824
NM_006283
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr1_+_178310605
|
0.718
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr10_+_104535887
|
0.718
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr19_+_34745452
|
0.711
|
NM_014686
|
KIAA0355
|
KIAA0355
|
|
chr7_-_28220342
|
0.700
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr6_-_3227776
|
0.690
|
NM_178012
|
TUBB2B
|
tubulin, beta 2B class IIb
|
|
chrX_-_64196268
|
0.688
|
NM_001178033
NM_018684
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chrX_+_107683015
|
0.686
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr7_+_106809405
|
0.671
|
NM_001244262
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr14_+_24836144
|
0.668
|
NM_001136022
NM_001198967
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr2_+_65283485
|
0.665
|
NM_015147
|
CEP68
|
centrosomal protein 68kDa
|
|
chr8_-_57123858
|
0.665
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr17_-_40575117
|
0.662
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chrX_-_64254592
|
0.655
|
NM_001178032
NM_001243804
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr2_+_12856813
|
0.649
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr6_+_36410543
|
0.648
|
NM_173562
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr11_-_117166247
|
0.644
|
NM_001207048
NM_001207049
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr14_-_53417808
|
0.641
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr2_+_109150807
|
0.631
|
NM_001193483
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr14_+_24838301
|
0.630
|
NM_001198966
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr21_+_47518012
|
0.620
|
NM_001849
NM_058174
NM_058175
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr17_+_57408907
|
0.620
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr3_-_48632592
|
0.618
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr10_+_120789227
|
0.617
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr1_+_33722158
|
0.616
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr21_-_28339405
|
0.611
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr14_+_55034329
|
0.610
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr6_-_111804413
|
0.609
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr7_+_12726910
|
0.607
|
NM_001195396
NM_005738
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr7_+_3340919
|
0.600
|
NM_152744
|
SDK1
|
sidekick cell adhesion molecule 1
|
|
chr14_+_77228132
|
0.598
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr5_-_134734915
|
0.595
|
NM_001040158
NM_138609
NM_138610
|
H2AFY
|
H2A histone family, member Y
|
|
chr10_+_12391471
|
0.594
|
NM_020397
NM_153498
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr16_+_23568833
|
0.592
|
NM_019116
|
UBFD1
|
ubiquitin family domain containing 1
|
|
chrX_-_18372777
|
0.585
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr9_-_110252045
|
0.585
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr8_+_26371461
|
0.584
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr22_-_44893903
|
0.584
|
NM_032287
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr1_-_155881163
|
0.584
|
NM_006912
|
RIT1
|
Ras-like without CAAX 1
|
|
chrX_-_92928566
|
0.577
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr2_+_109271267
|
0.571
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_-_219432955
|
0.570
|
NM_020935
|
USP37
|
ubiquitin specific peptidase 37
|
|
chr2_+_109065576
|
0.569
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr9_+_135037333
|
0.567
|
NM_032536
|
NTNG2
|
netrin G2
|
|
chr11_-_117186918
|
0.567
|
NM_012104
NM_138971
NM_138972
NM_138973
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr15_-_60919642
|
0.564
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_-_79103571
|
0.563
|
NM_014272
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7
|
|
chr2_+_109223600
|
0.562
|
NM_001193482
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr5_-_122372413
|
0.562
|
NM_000943
|
PPIC
|
peptidylprolyl isomerase C (cyclophilin C)
|
|
chr1_+_178062829
|
0.562
|
NM_170692
|
RASAL2
|
RAS protein activator like 2
|
|
chr9_+_35161950
|
0.560
|
NM_006377
|
UNC13B
|
unc-13 homolog B (C. elegans)
|
|
chr7_-_112579726
|
0.557
|
NM_152556
|
C7orf60
|
chromosome 7 open reading frame 60
|
|
chr17_+_45810609
|
0.556
|
NM_013351
|
TBX21
|
T-box 21
|
|
chr16_-_77468930
|
0.554
|
NM_199355
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18
|
|
chr19_-_10121041
|
0.549
|
NM_015719
|
COL5A3
|
collagen, type V, alpha 3
|
|
chr6_-_56112315
|
0.548
|
NM_030820
|
COL21A1
|
collagen, type XXI, alpha 1
|
|
chr3_+_72937168
|
0.546
|
NM_001080393
|
GXYLT2
|
glucoside xylosyltransferase 2
|
|
chr8_-_82024277
|
0.544
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr10_-_50747080
|
0.541
|
NM_000124
|
ERCC6
PGBD3
|
excision repair cross-complementing rodent repair deficiency, complementation group 6
piggyBac transposable element derived 3
|
|
chr20_+_46130600
|
0.537
|
NM_001174087
NM_001174088
NM_006534
NM_181659
|
NCOA3
|
nuclear receptor coactivator 3
|
|
chr19_-_38714781
|
0.536
|
NM_001135155
NM_004647
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr11_-_66335958
|
0.532
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr7_+_73442426
|
0.515
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chr8_+_26435339
|
0.514
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr3_-_64211111
|
0.513
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr17_+_21279667
|
0.513
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr17_+_67410825
|
0.511
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr1_+_31769826
|
0.509
|
NM_016505
|
ZCCHC17
|
zinc finger, CCHC domain containing 17
|
|
chr3_+_197687070
|
0.509
|
NM_001136049
NM_033029
|
LMLN
|
leishmanolysin-like (metallopeptidase M8 family)
|
|
chr20_-_3766905
|
0.505
|
NM_001810
|
CENPB
|
centromere protein B, 80kDa
|
|
chr1_-_225840660
|
0.504
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr2_-_220083699
|
0.502
|
NM_005689
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr13_-_53024730
|
0.500
|
NM_016075
|
VPS36
|
vacuolar protein sorting 36 homolog (S. cerevisiae)
|
|
chr2_-_61697846
|
0.497
|
NM_014709
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr1_-_150849211
|
0.496
|
NM_001197325
NM_001668
NM_178427
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator
|
|
chr17_+_14204326
|
0.493
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|