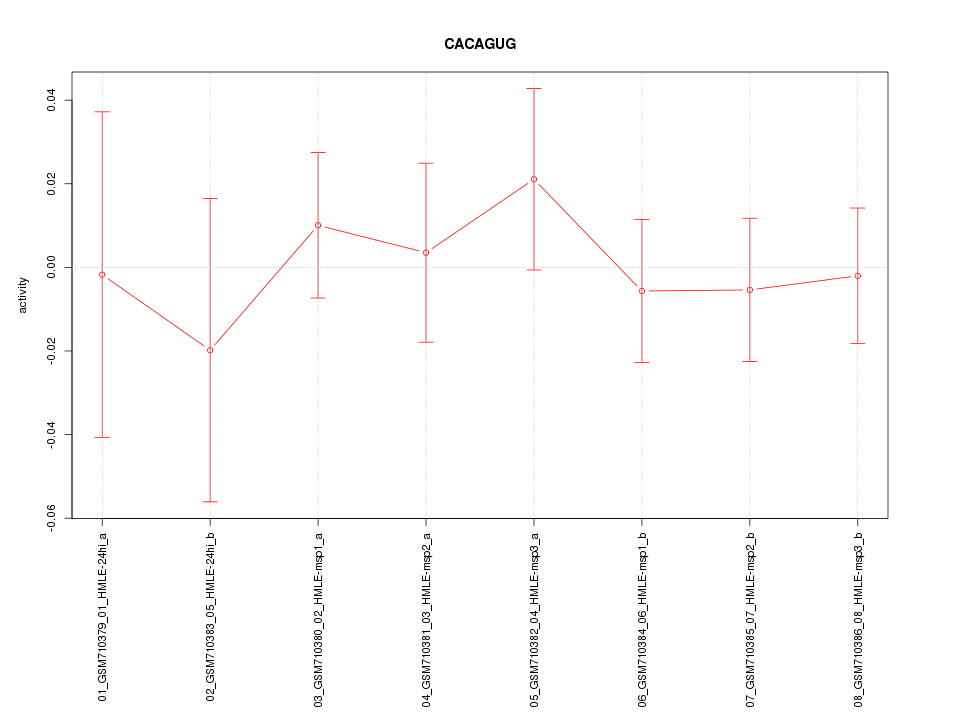
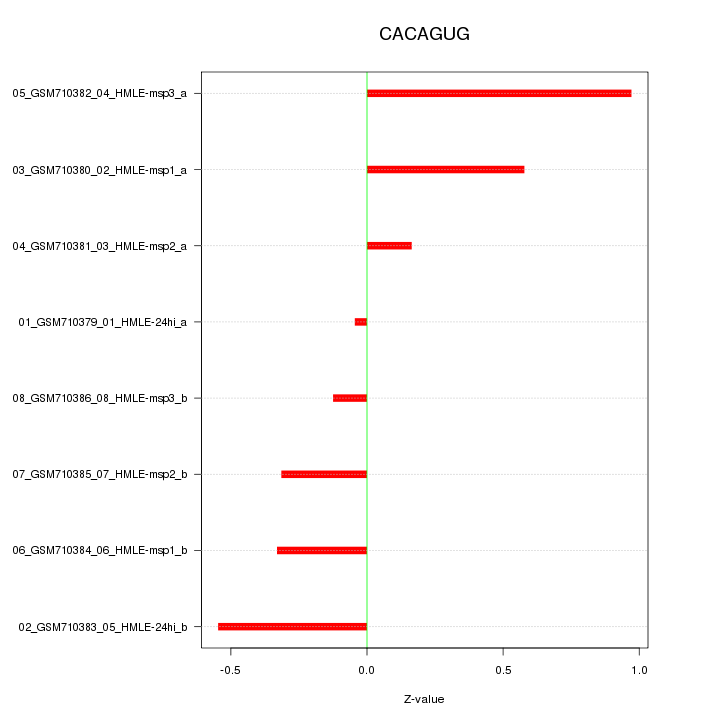
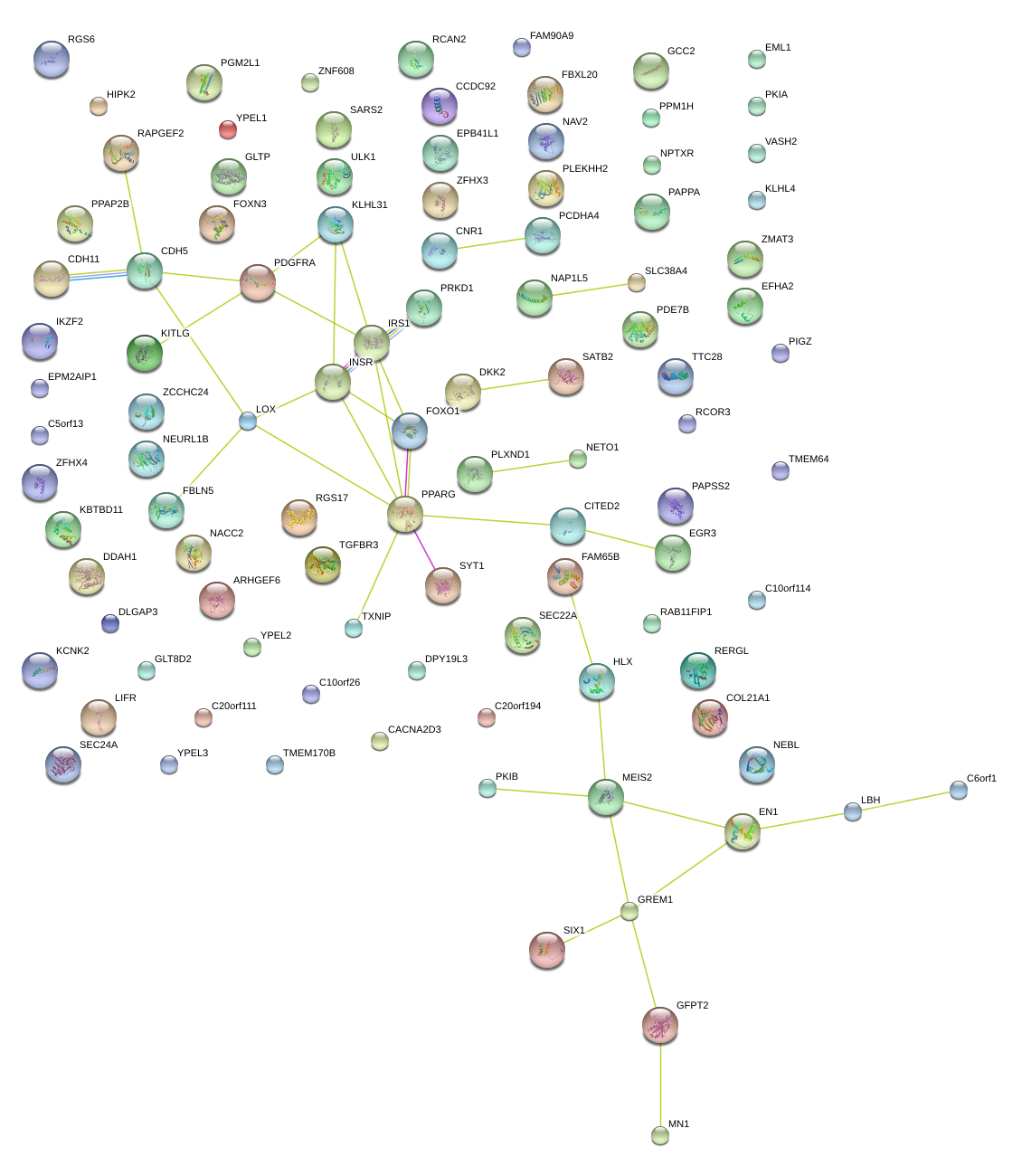
|
chr10_-_21463019
|
0.651
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr15_+_33010174
|
0.595
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr6_-_88875766
|
0.562
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr10_-_21186530
|
0.496
|
NM_006393
|
NEBL
|
nebulette
|
|
chr2_+_30454396
|
0.432
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr1_-_92371558
|
0.429
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr5_-_111312522
|
0.422
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_-_92351613
|
0.412
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr5_-_111093030
|
0.411
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093792
|
0.410
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093251
|
0.406
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111092918
|
0.402
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_-_57045235
|
0.401
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr5_-_179780314
|
0.395
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr1_+_221052735
|
0.355
|
NM_021958
|
HLX
|
H2.0-like homeobox
|
|
chr14_-_92414004
|
0.344
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr6_+_136172802
|
0.312
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr6_-_46293628
|
0.301
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr14_-_89883306
|
0.286
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr5_+_172068188
|
0.275
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr7_-_28998028
|
0.271
|
NM_014817
|
TRIL
|
TLR4 interactor with leucine-rich repeats
|
|
chr6_-_88854989
|
0.264
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr12_-_124457099
|
0.259
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr5_-_121412805
|
0.257
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr14_-_90085325
|
0.252
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr5_-_121414011
|
0.249
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr1_+_215256559
|
0.248
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr14_+_100259445
|
0.243
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr10_-_81205091
|
0.240
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr1_+_215178884
|
0.238
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr12_-_47219734
|
0.227
|
NM_001143824
NM_018018
|
SLC38A4
|
solute carrier family 38, member 4
|
|
chr8_+_1922030
|
0.226
|
NM_014867
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr6_-_24911194
|
0.221
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr19_+_32896515
|
0.218
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr8_+_79428335
|
0.216
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr9_+_118916069
|
0.213
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr12_+_79257772
|
0.212
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr19_+_32897021
|
0.210
|
NM_207325
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr14_-_30396811
|
0.199
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr12_-_63328663
|
0.197
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr6_-_53530505
|
0.194
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr2_-_214016332
|
0.194
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chrX_-_135863502
|
0.194
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr12_+_79439432
|
0.193
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr17_+_57408907
|
0.193
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr3_-_129325175
|
0.190
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr14_-_61115906
|
0.188
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr3_+_12330336
|
0.188
|
NM_138711
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr22_-_29075708
|
0.188
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr3_+_12392993
|
0.187
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr3_+_12329332
|
0.186
|
NM_005037
NM_138712
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr1_+_213123853
|
0.182
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr5_-_111091947
|
0.180
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr11_+_19734821
|
0.180
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr11_-_74109417
|
0.179
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr12_+_79258448
|
0.177
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr1_-_85930733
|
0.167
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr10_+_104503724
|
0.162
|
NM_001083913
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr4_+_55095263
|
0.159
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr20_+_34700257
|
0.159
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr20_+_34742656
|
0.158
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr8_-_91657898
|
0.158
|
NM_001008495
NM_001146273
|
TMEM64
|
transmembrane protein 64
|
|
chr2_+_109065576
|
0.157
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr11_+_19372270
|
0.157
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr2_+_43864433
|
0.154
|
NM_172069
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr12_-_88974094
|
0.153
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr8_+_16884745
|
0.153
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr7_-_139477437
|
0.152
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr16_-_30107492
|
0.152
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr1_-_86043932
|
0.149
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr9_-_138987096
|
0.147
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr6_-_139695349
|
0.147
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr8_+_77593506
|
0.146
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr6_+_11538486
|
0.145
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr3_+_119187782
|
0.145
|
NM_152305
|
POGLUT1
|
protein O-glucosyltransferase 1
|
|
chr5_-_38556682
|
0.145
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr6_-_139695784
|
0.143
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr3_+_54156691
|
0.138
|
NM_018398
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr10_-_21786205
|
0.134
|
NM_001010911
|
C10orf114
|
chromosome 10 open reading frame 114
|
|
chr16_-_65155881
|
0.129
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr10_+_104535887
|
0.128
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr8_-_37756971
|
0.126
|
NM_001002814
NM_025151
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr6_-_56112315
|
0.126
|
NM_030820
|
COL21A1
|
collagen, type XXI, alpha 1
|
|
chr22_-_28197469
|
0.124
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr8_-_22549901
|
0.124
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr1_-_35370983
|
0.124
|
NM_001080418
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr16_-_73082169
|
0.123
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr5_-_38595506
|
0.123
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr8_-_22550708
|
0.123
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr2_-_200335988
|
0.122
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr6_+_122931376
|
0.122
|
NM_032471
NM_181795
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta
|
|
chr1_+_211433285
|
0.119
|
NM_018254
|
RCOR3
|
REST corepressor 3
|
|
chr2_-_119605758
|
0.119
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr6_+_122793061
|
0.117
|
NM_181794
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta
|
|
chr3_-_196695608
|
0.115
|
NM_025163
|
PIGZ
|
phosphatidylinositol glycan anchor biosynthesis, class Z
|
|
chr8_-_22550057
|
0.115
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr16_+_66400524
|
0.113
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr12_-_110318178
|
0.112
|
NM_016433
|
GLTP
|
glycolipid transfer protein
|
|
chr17_-_37557851
|
0.112
|
NM_001184906
NM_032875
|
FBXL20
|
F-box and leucine-rich repeat protein 20
|
|
chr2_-_200322699
|
0.111
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr20_-_3388219
|
0.110
|
NM_001009984
|
C20orf194
|
chromosome 20 open reading frame 194
|
|
chr3_-_178789597
|
0.109
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr2_-_200329810
|
0.109
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr4_-_107957452
|
0.108
|
NM_014421
|
DKK2
|
dickkopf 2 homolog (Xenopus laevis)
|
|
chrX_+_86772714
|
0.108
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr22_-_39240016
|
0.107
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr1_+_145438437
|
0.107
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr20_-_42839423
|
0.106
|
NM_016470
|
C20orf111
|
chromosome 20 open reading frame 111
|
|
chr5_+_133984234
|
0.104
|
NM_001252231
NM_021982
|
SEC24A
|
SEC24 family, member A (S. cerevisiae)
|
|
chr15_-_37390372
|
0.102
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr2_-_227663454
|
0.102
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr3_+_122920773
|
0.102
|
NM_012430
|
SEC22A
|
SEC22 vesicle trafficking protein homolog A (S. cerevisiae)
|
|
chr5_-_124080773
|
0.101
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr12_+_132379262
|
0.100
|
NM_003565
|
ULK1
|
unc-51-like kinase 1 (C. elegans)
|
|
chr18_-_70534809
|
0.099
|
NM_001201465
NM_138966
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr2_-_214015053
|
0.099
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr6_-_153452383
|
0.098
|
NM_012419
|
RGS17
|
regulator of G-protein signaling 17
|
|
chr4_-_89618928
|
0.098
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr10_+_89419352
|
0.097
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr4_+_160188997
|
0.097
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr14_+_72398816
|
0.097
|
NM_001204423
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr13_-_41240607
|
0.097
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr19_-_7293876
|
0.096
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr7_-_27183225
|
0.096
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr3_+_43732358
|
0.095
|
NM_016006
|
ABHD5
|
abhydrolase domain containing 5
|
|
chr5_-_132299263
|
0.094
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr5_+_173315313
|
0.093
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr19_+_15218141
|
0.091
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr4_-_7941092
|
0.090
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr12_+_54422193
|
0.090
|
NM_004503
|
HOXC6
|
homeobox C6
|
|
chr16_+_2587951
|
0.090
|
NM_002613
NM_031268
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr6_+_126102278
|
0.089
|
NM_001199619
NM_001199620
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr10_-_90750955
|
0.089
|
NM_001141945
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr21_+_45285101
|
0.089
|
NM_020132
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr1_-_149889362
|
0.088
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr5_+_67511578
|
0.087
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr8_-_57123858
|
0.087
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr5_+_132149022
|
0.086
|
NM_175873
|
ANKRD43
|
ankyrin repeat domain 43
|
|
chr1_-_225840660
|
0.086
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr20_+_48599511
|
0.085
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr14_+_105266878
|
0.083
|
NM_001137601
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr5_+_162864571
|
0.083
|
NM_004060
NM_199246
|
CCNG1
|
cyclin G1
|
|
chr21_+_45345278
|
0.083
|
NM_001037553
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr16_+_66878281
|
0.083
|
NM_005182
|
CA7
|
carbonic anhydrase VII
|
|
chr1_-_51810693
|
0.082
|
NM_001144832
|
TTC39A
|
tetratricopeptide repeat domain 39A
|
|
chr2_+_24807345
|
0.082
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr15_-_60919642
|
0.082
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr1_+_23037329
|
0.082
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr5_+_67588395
|
0.082
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr12_+_103981050
|
0.082
|
NM_017564
|
STAB2
|
stabilin 2
|
|
chr5_+_67584251
|
0.081
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr12_+_131438451
|
0.080
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr3_-_124774735
|
0.080
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr7_-_112579726
|
0.080
|
NM_152556
|
C7orf60
|
chromosome 7 open reading frame 60
|
|
chr15_-_37392358
|
0.079
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr17_-_47439326
|
0.078
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr12_-_90049818
|
0.078
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr5_-_93447237
|
0.077
|
NM_001163417
NM_001163418
NM_032042
|
FAM172A
|
family with sequence similarity 172, member A
|
|
chr16_+_66878840
|
0.077
|
NM_001014435
|
CA7
|
carbonic anhydrase VII
|
|
chr17_-_2304217
|
0.077
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr5_+_67586466
|
0.076
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_-_33841142
|
0.075
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr15_-_37393364
|
0.074
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr9_-_130829598
|
0.073
|
NM_197956
|
NAIF1
|
nuclear apoptosis inducing factor 1
|
|
chr9_-_98269480
|
0.073
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr2_-_85555343
|
0.073
|
NM_001206840
NM_001206841
NM_001206844
NM_006464
|
TGOLN2
|
trans-golgi network protein 2
|
|
chr3_-_87040246
|
0.073
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chrX_+_9754465
|
0.073
|
NM_001649
|
SHROOM2
|
shroom family member 2
|
|
chr12_-_49182718
|
0.072
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr2_+_210636700
|
0.072
|
NM_032504
NM_182587
|
UNC80
|
unc-80 homolog (C. elegans)
|
|
chrX_+_108780235
|
0.072
|
NM_001242618
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr11_+_12695851
|
0.071
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr3_-_141868208
|
0.071
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr20_-_3149070
|
0.070
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr1_+_211432693
|
0.070
|
NM_001136223
NM_001136224
NM_001136225
|
RCOR3
|
REST corepressor 3
|
|
chr1_+_101361589
|
0.070
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr6_+_126111782
|
0.070
|
NM_001122842
NM_001199621
NM_181782
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr15_+_91447419
|
0.069
|
NM_006122
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2
|
|
chr1_-_47134039
|
0.069
|
NM_001042546
NM_022745
|
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1
|
|
chr1_+_27561006
|
0.069
|
NM_015023
|
WDTC1
|
WD and tetratricopeptide repeats 1
|
|
chrX_+_51486479
|
0.068
|
NM_018094
|
GSPT2
|
G1 to S phase transition 2
|
|
chr15_-_59949536
|
0.068
|
NM_004492
|
GTF2A2
|
general transcription factor IIA, 2, 12kDa
|
|
chr15_-_60884615
|
0.067
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr10_-_91403630
|
0.066
|
NM_138316
NM_148978
|
PANK1
|
pantothenate kinase 1
|
|
chr10_-_94050799
|
0.066
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr9_-_98269686
|
0.066
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr5_-_64920149
|
0.065
|
NM_001656
NM_033227
NM_033228
|
TRIM23
|
tripartite motif containing 23
|
|
chr10_-_91405320
|
0.064
|
NM_148977
|
PANK1
|
pantothenate kinase 1
|
|
chr9_+_127020462
|
0.064
|
NM_001166168
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chrX_+_108779009
|
0.064
|
NM_018698
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr9_+_127054246
|
0.064
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr6_+_170102236
|
0.063
|
NM_001029863
|
C6orf120
|
chromosome 6 open reading frame 120
|
|
chr20_-_62610990
|
0.063
|
NM_080621
|
SAMD10
|
sterile alpha motif domain containing 10
|
|
chr3_-_48594209
|
0.063
|
NM_004567
|
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
|
|
chr4_-_13546037
|
0.063
|
NM_001189
|
NKX3-2
|
NK3 homeobox 2
|
|
chr1_-_37499708
|
0.063
|
NM_000831
|
GRIK3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr14_+_72399149
|
0.063
|
NM_001204424
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr15_-_61521478
|
0.061
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr3_+_156008775
|
0.061
|
NM_172159
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr20_-_20693122
|
0.061
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|