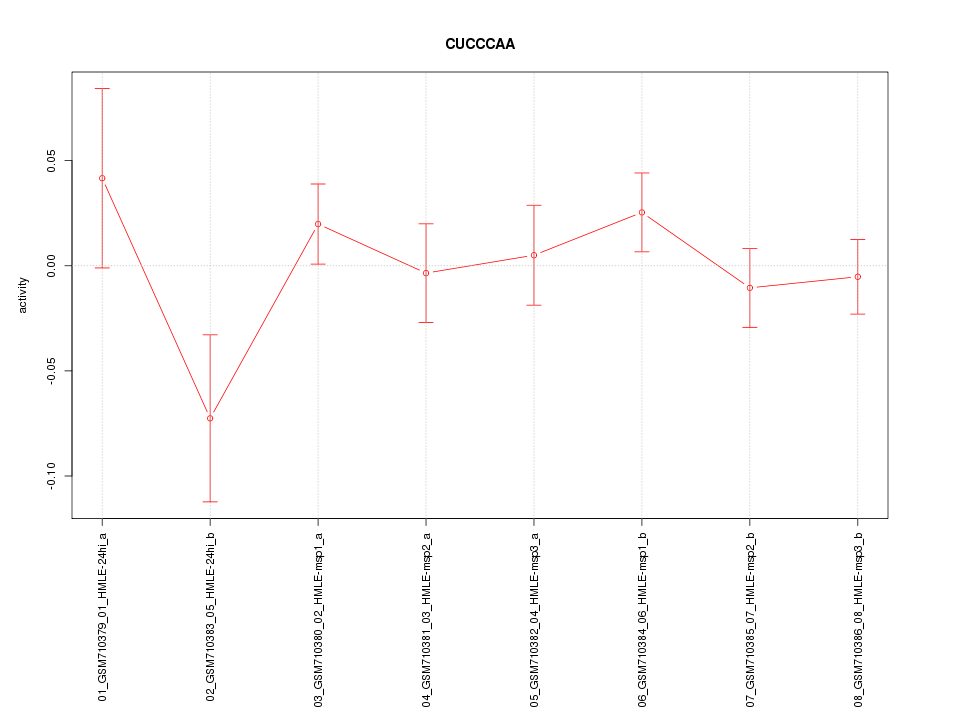
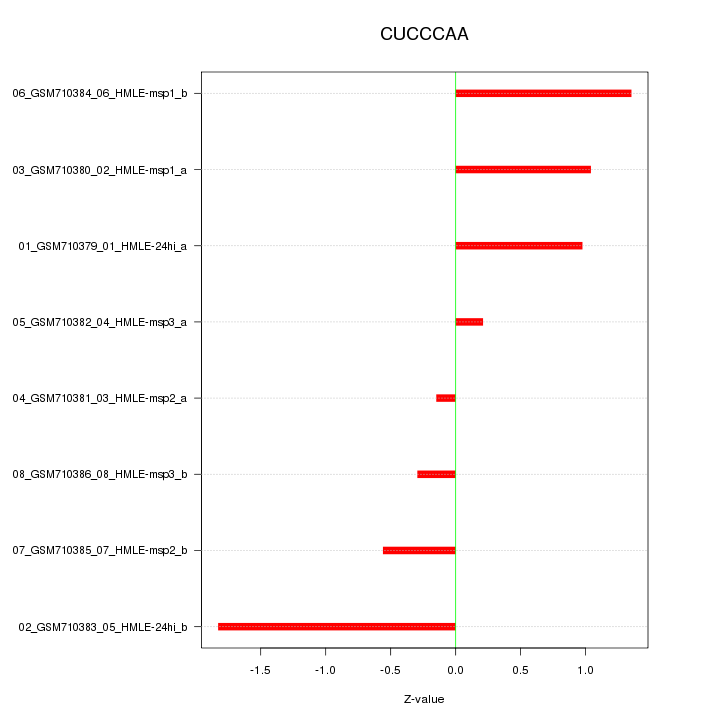
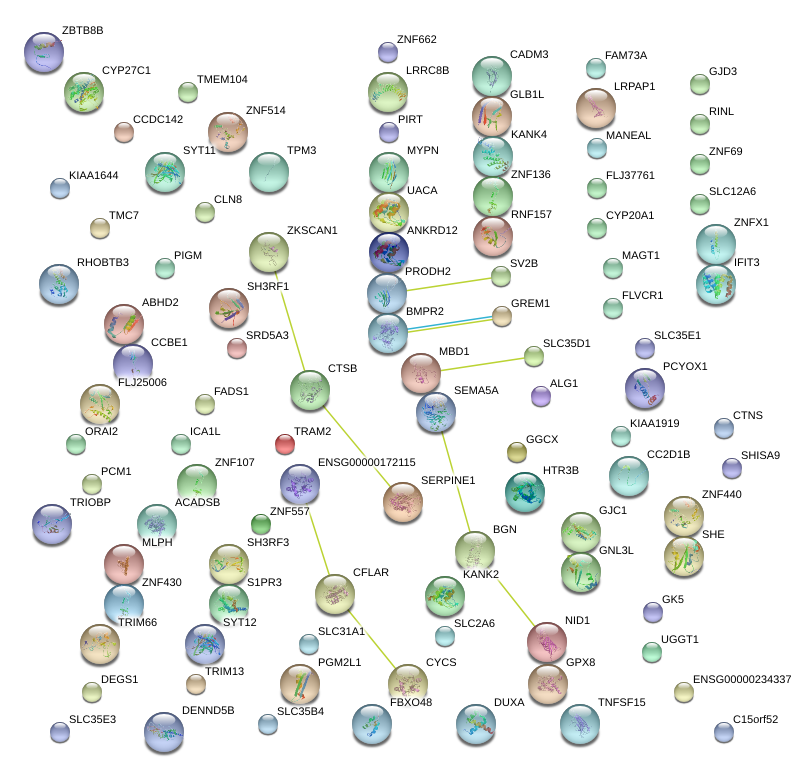
|
chr15_+_33010174
|
0.424
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr1_+_228353428
|
0.403
|
NM_001010867
|
IBA57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae)
|
|
chr2_+_204103163
|
0.399
|
NM_177538
|
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1
|
|
chr5_-_9546157
|
0.371
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr8_+_1711869
|
0.354
|
NM_018941
|
CLN8
|
ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation)
|
|
chr15_+_91643429
|
0.340
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr17_-_42907606
|
0.333
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr9_-_136344182
|
0.332
|
NM_001145099
NM_017585
|
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6
|
|
chr22_-_39052579
|
0.332
|
NM_001013647
|
LOC646851
|
putative uncharacterized protein LOC388900
|
|
chr2_-_127963342
|
0.324
|
NM_001001665
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1
|
|
chr7_-_25164902
|
0.324
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr17_-_42908178
|
0.317
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr2_+_201997687
|
0.314
|
NM_001202518
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr19_-_39368889
|
0.313
|
NM_001195833
NM_198445
|
RINL
|
Ras and Rab interactor-like
|
|
chr7_+_100770378
|
0.304
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr2_+_202004997
|
0.294
|
NM_001202515
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr1_+_159141376
|
0.282
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr1_+_213031596
|
0.277
|
NM_014053
|
FLVCR1
|
feline leukemia virus subgroup C cellular receptor 1
|
|
chr2_+_109745996
|
0.271
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr17_+_3539761
|
0.267
|
NM_001031681
NM_004937
|
CTNS
|
cystinosin, lysosomal cystine transporter
|
|
chr15_-_40633122
|
0.261
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr22_+_38092994
|
0.260
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr3_-_141944441
|
0.259
|
NM_001039547
|
GK5
|
glycerol kinase 5 (putative)
|
|
chr12_+_69139917
|
0.249
|
NM_018656
|
SLC35E3
|
solute carrier family 35, member E3
|
|
chr1_+_89990396
|
0.242
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr16_+_5121809
|
0.242
|
NM_019109
|
ALG1
|
asparagine-linked glycosylation 1, beta-1,4-mannosyltransferase homolog (S. cerevisiae)
|
|
chr2_+_201983268
|
0.239
|
NM_001127183
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr2_+_70485204
|
0.236
|
NM_016297
|
PCYOX1
|
prenylcysteine oxidase 1
|
|
chr11_-_61584451
|
0.233
|
NM_013402
|
FADS1
|
fatty acid desaturase 1
|
|
chr4_-_3534153
|
0.231
|
NM_002337
|
LRPAP1
|
low density lipoprotein receptor-related protein associated protein 1
|
|
chr16_+_18995255
|
0.222
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr6_+_111580481
|
0.222
|
NM_153369
|
KIAA1919
|
KIAA1919
|
|
chr7_-_134001749
|
0.220
|
NM_032826
|
SLC35B4
|
solute carrier family 35, member B4
|
|
chr17_-_10741398
|
0.216
|
NM_001101387
|
PIRT
|
phosphoinositide-interacting regulator of transient receptor potential channels
|
|
chr16_+_18995508
|
0.214
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr7_+_102073953
|
0.211
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr1_+_90024271
|
0.206
|
NM_015350
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr1_-_160001723
|
0.204
|
NM_145167
|
PIGM
|
phosphatidylinositol glycan anchor biosynthesis, class M
|
|
chr22_-_44708730
|
0.204
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr17_-_74236272
|
0.202
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr1_-_236228476
|
0.194
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr10_+_91092238
|
0.193
|
NM_001031683
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr18_-_57364643
|
0.192
|
NM_133459
|
CCBE1
|
collagen and calcium binding EGF domains 1
|
|
chr1_-_62785067
|
0.192
|
NM_181712
|
KANK4
|
KN motif and ankyrin repeat domains 4
|
|
chr11_-_74109417
|
0.189
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr2_-_74710356
|
0.187
|
NM_032779
|
CCDC142
|
coiled-coil domain containing 142
|
|
chr5_+_95066628
|
0.184
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr2_+_238395863
|
0.181
|
NM_001042467
NM_024101
|
MLPH
|
melanophilin
|
|
chr2_-_203736223
|
0.181
|
NM_138468
|
ICA1L
|
islet cell autoantigen 1,69kDa-like
|
|
chrX_+_54556633
|
0.181
|
NM_001184819
NM_019067
|
GNL3L
|
guanine nucleotide binding protein-like 3 (nucleolar)-like
|
|
chr4_+_56212381
|
0.180
|
NM_024592
|
SRD5A3
|
steroid 5 alpha-reductase 3
|
|
chr2_-_85788644
|
0.177
|
NM_000821
NM_001142269
|
GGCX
|
gamma-glutamyl carboxylase
|
|
chr8_+_17780351
|
0.176
|
NM_006197
|
PCM1
|
pericentriolar material 1
|
|
chr19_+_7069454
|
0.174
|
NM_001044387
NM_001044388
NM_024341
|
ZNF557
|
zinc finger protein 557
|
|
chr19_-_11308184
|
0.174
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr11_+_113775517
|
0.173
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr2_+_201980873
|
0.171
|
NM_001127184
NM_001202516
NM_001202517
NM_001202519
NM_003879
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr2_-_220110116
|
0.171
|
NM_024506
|
GLB1L
|
galactosidase, beta 1-like
|
|
chr11_-_8680382
|
0.170
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chr19_-_57678855
|
0.170
|
NM_001012729
|
DUXA
|
double homeobox A
|
|
chr2_+_128848752
|
0.169
|
NM_020120
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr20_-_47894591
|
0.166
|
NM_021035
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chrX_+_152760346
|
0.166
|
NM_001711
|
BGN
|
biglycan
|
|
chrX_-_77150907
|
0.165
|
NM_032121
|
MAGT1
|
magnesium transporter 1
|
|
chr10_+_69869249
|
0.165
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr1_-_154474524
|
0.159
|
NM_001010846
|
SHE
|
Src homology 2 domain containing E
|
|
chr10_+_91087601
|
0.159
|
NM_001549
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr15_+_89631380
|
0.159
|
NM_007011
NM_152924
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr19_-_11305186
|
0.158
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr7_+_64126481
|
0.156
|
NM_001013746
NM_016220
|
ZNF107
|
zinc finger protein 107
|
|
chr19_-_36342894
|
0.156
|
NM_004646
|
NPHS1
|
nephrosis 1, congenital, Finnish type (nephrin)
|
|
chr18_+_9136742
|
0.156
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr1_+_224370914
|
0.155
|
NM_003676
|
DEGS1
|
delta(4)-desaturase, sphingolipid 1
|
|
chr11_-_1771823
|
0.155
|
NM_001170820
|
IFITM10
|
interferon induced transmembrane protein 10
|
|
chr1_+_32930638
|
0.155
|
NM_001145720
|
ZBTB8B
|
zinc finger and BTB domain containing 8B
|
|
chr1_+_78245306
|
0.154
|
NM_198549
|
FAM73A
|
family with sequence similarity 73, member A
|
|
chr1_-_154164551
|
0.152
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr17_+_72772621
|
0.151
|
NM_017728
|
TMEM104
|
transmembrane protein 104
|
|
chr18_+_9137560
|
0.150
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr13_+_50571137
|
0.149
|
NM_001007278
NM_005798
NM_052811
NM_213590
|
TRIM13
|
tripartite motif containing 13
|
|
chr19_+_21203425
|
0.147
|
NM_001172671
NM_025189
|
ZNF430
|
zinc finger protein 430
|
|
chr9_+_91606349
|
0.145
|
NM_005226
|
S1PR3
|
sphingosine-1-phosphate receptor 3
|
|
chr1_+_155829259
|
0.144
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr6_-_52441815
|
0.142
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr1_+_38259458
|
0.142
|
NM_001031740
NM_001113482
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr3_+_42947649
|
0.142
|
NM_001134656
|
ZNF662
|
zinc finger protein 662
|
|
chr8_-_11725628
|
0.139
|
NM_001908
NM_147780
NM_147781
NM_147782
NM_147783
|
CTSB
|
cathepsin B
|
|
chr7_+_99613174
|
0.138
|
NM_003439
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1
|
|
chr19_-_16683188
|
0.137
|
NM_024881
|
SLC35E1
|
solute carrier family 35, member E1
|
|
chr19_+_12273866
|
0.136
|
NM_003437
|
ZNF136
|
zinc finger protein 136
|
|
chr2_+_203241044
|
0.135
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chr1_+_38261079
|
0.135
|
NM_152496
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr9_+_115983807
|
0.134
|
NM_001859
|
SLC31A1
|
solute carrier family 31 (copper transporters), member 1
|
|
chr15_-_70994608
|
0.134
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr10_+_124768401
|
0.132
|
NM_001609
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chr20_+_54933982
|
0.132
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr1_-_67520028
|
0.131
|
NM_015139
|
SLC35D1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1
|
|
chr3_+_42947401
|
0.131
|
NM_207404
|
ZNF662
|
zinc finger protein 662
|
|
chr2_-_68694389
|
0.130
|
NM_001024680
|
FBXO48
|
F-box protein 48
|
|
chr2_-_95825262
|
0.130
|
NM_032788
|
ZNF514
|
zinc finger protein 514
|
|
chr15_-_34630203
|
0.130
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr19_+_11925077
|
0.130
|
NM_152357
|
ZNF440
|
zinc finger protein 440
|
|
chr16_+_12995454
|
0.129
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr12_-_89918531
|
0.129
|
NM_003774
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4)
|
|
chr12_-_31743822
|
0.127
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr17_-_26941178
|
0.127
|
NM_001174103
|
SGK494
|
uncharacterized serine/threonine-protein kinase SgK494
|
|
chr5_+_54455945
|
0.125
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr1_-_52831817
|
0.125
|
NM_032449
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr9_-_117568292
|
0.125
|
NM_005118
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr19_-_52598965
|
0.124
|
NM_001136499
|
ZNF841
|
zinc finger protein 841
|
|
chr6_-_7313352
|
0.124
|
NM_003144
|
SSR1
|
signal sequence receptor, alpha
|
|
chr11_+_44587140
|
0.124
|
NM_001024844
NM_002231
|
CD82
|
CD82 molecule
|
|
chr16_-_23724820
|
0.123
|
NM_033266
|
ERN2
|
endoplasmic reticulum to nucleus signaling 2
|
|
chr3_+_121286777
|
0.123
|
NM_001012659
|
ARGFX
|
arginine-fifty homeobox
|
|
chr2_-_98206253
|
0.122
|
NM_025190
|
ANKRD36B
|
ankyrin repeat domain 36B
|
|
chr5_-_137368778
|
0.122
|
NM_001101800
NM_001101801
NM_016603
|
FAM13B
|
family with sequence similarity 13, member B
|
|
chr19_+_58193336
|
0.122
|
NM_138347
|
ZNF551
|
zinc finger protein 551
|
|
chr8_-_134584012
|
0.122
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr16_+_89787383
|
0.122
|
NM_152287
|
ZNF276
|
zinc finger protein 276
|
|
chr22_+_44319618
|
0.122
|
NM_025225
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr7_-_102789334
|
0.122
|
NM_001122838
NM_198990
|
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D
|
|
chr6_-_38607923
|
0.121
|
NM_001099272
NM_052893
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chrX_+_119737743
|
0.121
|
NM_014060
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr3_+_49449638
|
0.120
|
NM_022171
|
TCTA
|
T-cell leukemia translocation altered gene
|
|
chrX_+_16964730
|
0.120
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr15_-_42565473
|
0.119
|
NM_001110503
NM_015497
|
TMEM87A
|
transmembrane protein 87A
|
|
chr5_-_157002739
|
0.119
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr1_+_19638739
|
0.118
|
NM_001040125
NM_001040126
NM_017765
|
PQLC2
|
PQ loop repeat containing 2
|
|
chr11_-_63536112
|
0.117
|
NM_001144936
|
C11orf95
|
chromosome 11 open reading frame 95
|
|
chr12_-_69326946
|
0.117
|
NM_001005502
NM_198320
|
CPM
|
carboxypeptidase M
|
|
chr3_-_9595412
|
0.116
|
NM_198560
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4
|
|
chr15_-_34610928
|
0.116
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr11_-_108368918
|
0.116
|
NM_153705
|
KDELC2
|
KDEL (Lys-Asp-Glu-Leu) containing 2
|
|
chr1_-_1293907
|
0.116
|
NM_032348
|
MXRA8
|
matrix-remodelling associated 8
|
|
chr17_+_38599651
|
0.115
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr5_-_145562235
|
0.115
|
NM_020117
|
LARS
|
leucyl-tRNA synthetase
|
|
chr8_+_98656329
|
0.114
|
NM_178812
|
MTDH
|
metadherin
|
|
chr15_-_71055745
|
0.113
|
NM_018003
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr12_-_53574379
|
0.113
|
NM_015989
|
CSAD
|
cysteine sulfinic acid decarboxylase
|
|
chr22_+_38142230
|
0.113
|
NM_007032
NM_138632
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr2_-_85555343
|
0.112
|
NM_001206840
NM_001206841
NM_001206844
NM_006464
|
TGOLN2
|
trans-golgi network protein 2
|
|
chr12_+_65672682
|
0.112
|
NM_001193461
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr17_+_72462521
|
0.112
|
NM_007261
|
CD300A
|
CD300a molecule
|
|
chr5_+_134209426
|
0.112
|
NM_024715
|
TXNDC15
|
thioredoxin domain containing 15
|
|
chr7_-_72993007
|
0.112
|
NM_012453
|
TBL2
|
transducin (beta)-like 2
|
|
chr3_+_11196213
|
0.111
|
NM_001098212
|
HRH1
|
histamine receptor H1
|
|
chr1_+_180601121
|
0.110
|
NM_001135669
NM_004736
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr5_+_149340287
|
0.110
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr2_-_175547471
|
0.109
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr6_-_108395939
|
0.109
|
NM_014028
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr1_-_1624180
|
0.109
|
NM_001110781
|
SLC35E2B
|
solute carrier family 35, member E2B
|
|
chr10_+_91174303
|
0.109
|
NM_012420
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5
|
|
chr19_-_14889347
|
0.108
|
NM_013447
NM_152916
NM_152917
NM_152918
NM_152919
NM_152920
NM_152921
|
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2
|
|
chr4_+_123747815
|
0.108
|
NM_002006
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr9_+_5450455
|
0.108
|
NM_014143
|
CD274
|
CD274 molecule
|
|
chr3_+_11178778
|
0.108
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr3_+_11267666
|
0.108
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr19_+_53935222
|
0.107
|
NM_001008401
|
ZNF761
|
zinc finger protein 761
|
|
chr19_-_40562107
|
0.106
|
NM_001005851
|
ZNF780B
|
zinc finger protein 780B
|
|
chr19_-_58238895
|
0.106
|
NM_024833
|
ZNF671
|
zinc finger protein 671
|
|
chr3_+_43732358
|
0.106
|
NM_016006
|
ABHD5
|
abhydrolase domain containing 5
|
|
chr12_-_108733075
|
0.106
|
NM_001142343
NM_001142344
NM_004072
|
CMKLR1
|
chemokine-like receptor 1
|
|
chr3_-_49314281
|
0.106
|
NM_198562
|
C3orf62
|
chromosome 3 open reading frame 62
|
|
chr11_-_777470
|
0.105
|
NM_182612
|
PDDC1
|
Parkinson disease 7 domain containing 1
|
|
chr15_+_42867708
|
0.105
|
NM_020759
|
STARD9
|
StAR-related lipid transfer (START) domain containing 9
|
|
chr15_-_34629941
|
0.104
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr12_+_32654976
|
0.104
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr6_-_135271206
|
0.104
|
NM_001193480
NM_022568
NM_170771
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1
|
|
chr19_+_48972464
|
0.103
|
NM_004228
NM_017457
|
CYTH2
|
cytohesin 2
|
|
chr19_-_53662279
|
0.103
|
NM_001172674
NM_001172675
NM_032584
|
ZNF347
|
zinc finger protein 347
|
|
chr4_+_40058507
|
0.103
|
NM_018177
|
N4BP2
|
NEDD4 binding protein 2
|
|
chr3_+_11294384
|
0.103
|
NM_000861
|
HRH1
|
histamine receptor H1
|
|
chr11_-_30607762
|
0.103
|
NM_001145399
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr11_-_33744272
|
0.102
|
NM_001127223
NM_001127225
NM_001127226
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr2_-_175499275
|
0.102
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr11_-_33744497
|
0.101
|
NM_001127227
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr10_+_94352809
|
0.101
|
NM_004523
|
KIF11
|
kinesin family member 11
|
|
chr19_+_58258160
|
0.101
|
NM_173632
|
ZNF776
|
zinc finger protein 776
|
|
chr17_+_74732629
|
0.101
|
NM_001242534
|
MFSD11
|
major facilitator superfamily domain containing 11
|
|
chr4_+_155665150
|
0.101
|
NM_004744
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase)
|
|
chr14_+_74111577
|
0.100
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chr2_+_17935379
|
0.100
|
NM_001130009
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr15_-_34629044
|
0.100
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_+_110693131
|
0.100
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr13_-_73356070
|
0.100
|
NM_001128226
NM_014953
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr19_+_58740069
|
0.099
|
NM_014480
|
ZNF544
|
zinc finger protein 544
|
|
chr2_-_207082626
|
0.099
|
NM_001098199
NM_005279
|
GPR1
|
G protein-coupled receptor 1
|
|
chr1_-_113161724
|
0.099
|
NM_017744
NM_138727
NM_138728
NM_138729
|
ST7L
|
suppression of tumorigenicity 7 like
|
|
chr19_+_14800612
|
0.099
|
NM_032433
|
ZNF333
|
zinc finger protein 333
|
|
chr1_-_208417635
|
0.099
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr15_+_74466024
|
0.098
|
NM_005545
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr4_-_74904329
|
0.098
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr4_+_15704572
|
0.098
|
NM_004334
|
BST1
|
bone marrow stromal cell antigen 1
|
|
chr17_+_74733468
|
0.098
|
NM_001242532
NM_001242533
NM_001242535
NM_001242536
NM_001242537
NM_024311
|
MFSD11
|
major facilitator superfamily domain containing 11
|
|
chr7_-_55930444
|
0.098
|
NM_207366
|
SEPT14
|
septin 14
|
|
chr9_-_33264458
|
0.098
|
NM_001172415
NM_004323
|
BAG1
|
BCL2-associated athanogene
|
|
chr16_+_89787951
|
0.098
|
NM_001113525
|
ZNF276
|
zinc finger protein 276
|
|
chr17_-_8301143
|
0.098
|
NM_001146684
|
RNF222
|
ring finger protein 222
|
|
chr12_-_69357019
|
0.097
|
NM_001874
|
CPM
|
carboxypeptidase M
|
|
chr3_+_105085553
|
0.097
|
NM_001243280
NM_001243281
NM_001243283
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|