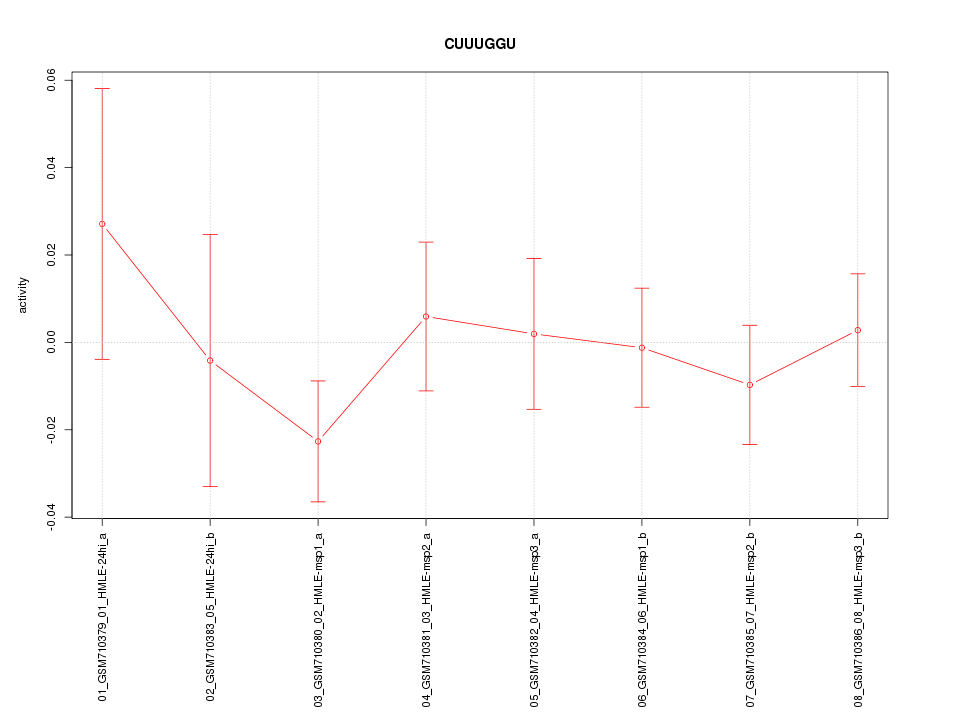
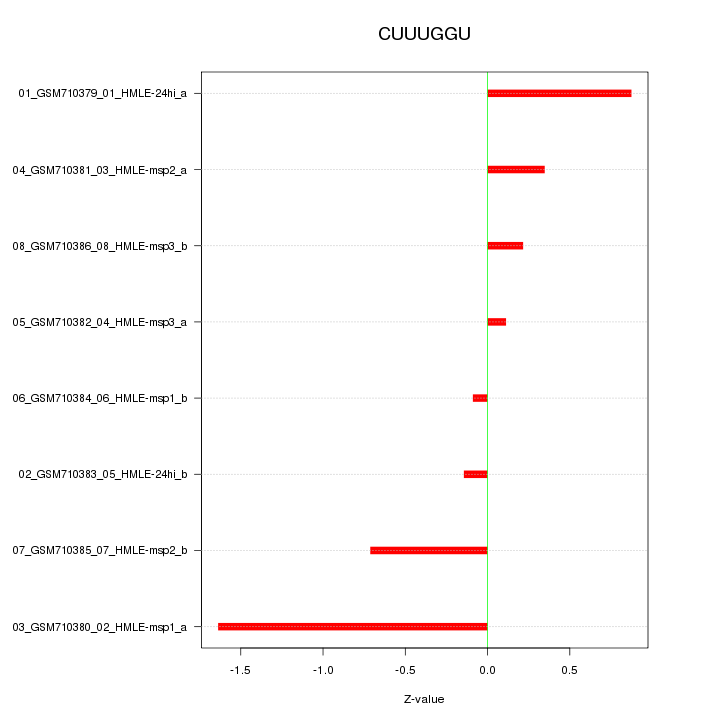
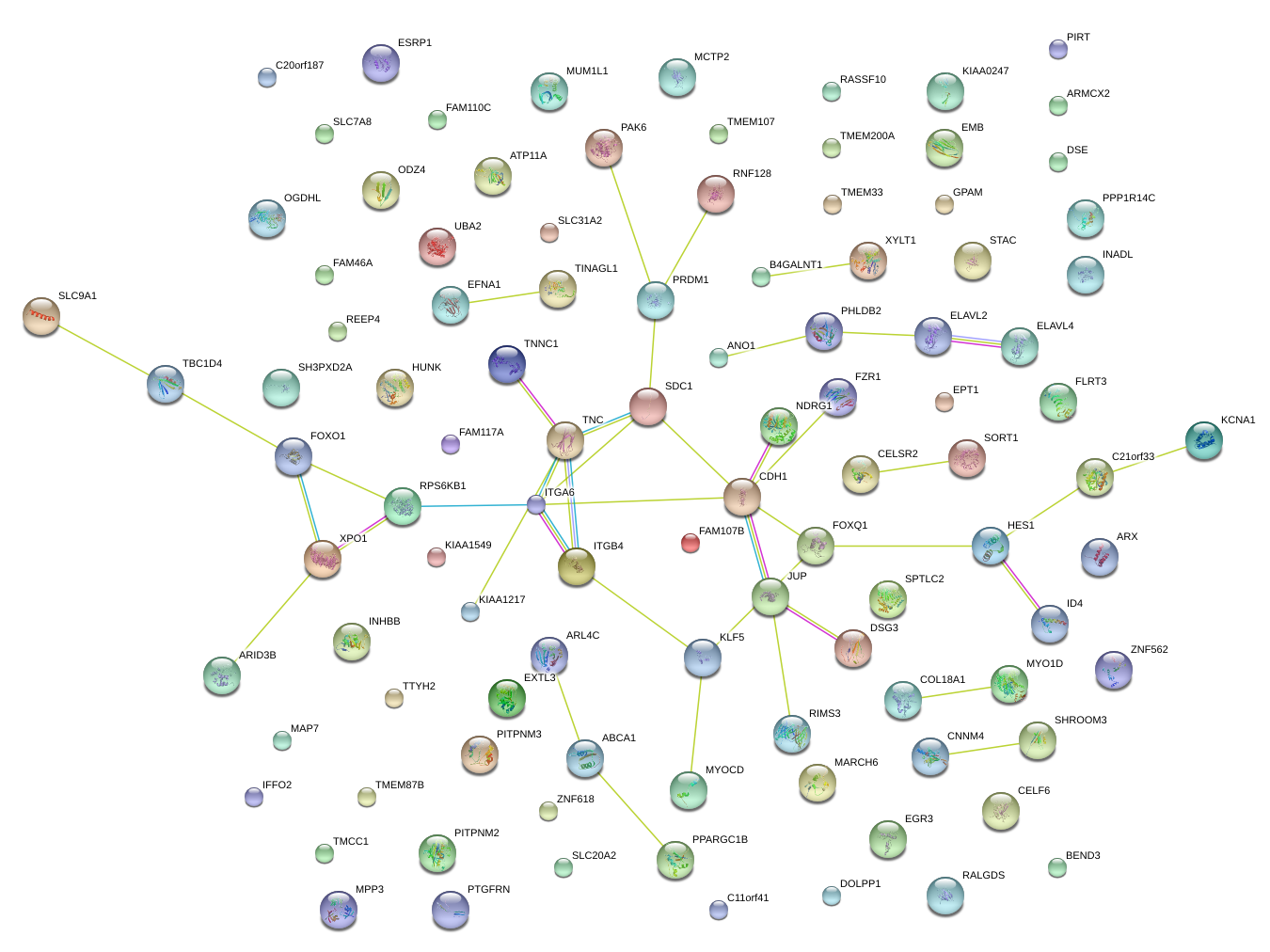
|
chr11_+_69924407
|
0.706
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr16_+_68771192
|
0.689
|
NM_004360
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr2_-_46384
|
0.498
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr1_+_155100304
|
0.487
|
NM_004428
NM_182685
|
EFNA1
|
ephrin-A1
|
|
chr6_+_130758261
|
0.448
|
NM_052913
|
TMEM200A
|
transmembrane protein 200A
|
|
chr4_+_77356252
|
0.439
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr6_-_136847743
|
0.437
|
NM_001198614
NM_001198618
NM_001198619
|
MAP7
|
microtubule-associated protein 7
|
|
chr6_-_136871956
|
0.434
|
NM_001198616
NM_001198617
NM_003980
|
MAP7
|
microtubule-associated protein 7
|
|
chr6_-_136788012
|
0.422
|
NM_001198615
|
MAP7
|
microtubule-associated protein 7
|
|
chr9_+_115913227
|
0.410
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr6_-_136847078
|
0.409
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr15_+_40509628
|
0.394
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr11_-_79151694
|
0.391
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr15_+_40532034
|
0.372
|
NM_020168
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr6_+_150464156
|
0.367
|
NM_030949
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C
|
|
chr1_+_117452359
|
0.336
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr6_+_106534188
|
0.334
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr14_-_23652840
|
0.333
|
NM_012244
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr1_+_109792640
|
0.327
|
NM_001408
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr9_-_117880411
|
0.313
|
NM_002160
|
TNC
|
tenascin C
|
|
chr9_-_107690526
|
0.312
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr14_-_23623493
|
0.309
|
NM_182728
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr6_+_106546736
|
0.302
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr8_+_95653340
|
0.300
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr10_+_23983674
|
0.296
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chrX_+_105412297
|
0.289
|
NM_001171020
NM_152423
|
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1
|
|
chr2_-_20425166
|
0.281
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr20_-_14318200
|
0.279
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr18_+_29027731
|
0.278
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr9_-_23826062
|
0.275
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr17_-_39942937
|
0.271
|
NM_002230
NM_021991
|
JUP
|
junction plakoglobin
|
|
chr2_-_235405692
|
0.270
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr15_-_72612497
|
0.268
|
NM_052840
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr2_+_173292306
|
0.267
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr15_-_72612247
|
0.266
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr9_-_23821842
|
0.264
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr1_+_32042085
|
0.263
|
NM_001204414
NM_022164
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chrX_-_100914816
|
0.263
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr2_-_20424627
|
0.262
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr11_+_33563772
|
0.261
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chrX_+_105937067
|
0.255
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr9_-_23821477
|
0.252
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr8_-_22550708
|
0.251
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr1_+_32043745
|
0.249
|
NM_001204415
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr11_+_13030879
|
0.245
|
NM_001080521
|
RASSF10
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 10
|
|
chr17_+_73717515
|
0.241
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr8_-_134309448
|
0.239
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr8_-_22550057
|
0.237
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chrX_+_105969875
|
0.237
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr10_-_105614952
|
0.237
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr8_-_22549901
|
0.235
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr17_-_31203889
|
0.226
|
NM_015194
|
MYO1D
|
myosin ID
|
|
chr9_-_135996287
|
0.221
|
NM_006266
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr6_+_1312674
|
0.221
|
NM_033260
|
FOXQ1
|
forkhead box Q1
|
|
chr21_+_46875402
|
0.219
|
NM_030582
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr6_+_116692098
|
0.219
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr21_+_46825001
|
0.219
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr3_+_36421944
|
0.218
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr9_+_116638544
|
0.212
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr2_+_112812799
|
0.211
|
NM_032824
|
TMEM87B
|
transmembrane protein 87B
|
|
chr12_+_5019048
|
0.207
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr12_-_123594974
|
0.205
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr2_+_97426571
|
0.202
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr8_-_42397048
|
0.201
|
NM_006749
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2
|
|
chr17_+_73720775
|
0.200
|
NM_001005619
|
ITGB4
|
integrin, beta 4
|
|
chr17_-_47841517
|
0.199
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr15_+_74833517
|
0.199
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr15_-_72598655
|
0.196
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr10_-_50970321
|
0.195
|
NM_001143996
NM_001143997
NM_018245
|
OGDHL
|
oxoglutarate dehydrogenase-like
|
|
chr17_-_10741398
|
0.195
|
NM_001101387
|
PIRT
|
phosphoinositide-interacting regulator of transient receptor potential channels
|
|
chrX_-_25034064
|
0.194
|
NM_139058
|
ARX
|
aristaless related homeobox
|
|
chr10_-_14816895
|
0.192
|
NM_031453
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr9_-_136024522
|
0.191
|
NM_001042368
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr4_+_41937105
|
0.189
|
NM_018126
|
TMEM33
|
transmembrane protein 33
|
|
chr21_+_33245304
|
0.187
|
NM_014586
|
HUNK
|
hormonally up-regulated Neu-associated kinase
|
|
chr8_+_28559143
|
0.187
|
NM_001440
|
EXTL3
|
exostoses (multiple)-like 3
|
|
chr8_-_21998994
|
0.186
|
NM_025232
|
REEP4
|
receptor accessory protein 4
|
|
chr6_-_82462374
|
0.183
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr12_-_58026846
|
0.180
|
NM_001478
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1
|
|
chr1_-_41131323
|
0.180
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr3_+_111451326
|
0.179
|
NM_001134437
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr17_-_41910439
|
0.178
|
NM_001932
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3)
|
|
chr3_+_193853930
|
0.178
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr1_-_27481400
|
0.177
|
NM_003047
|
SLC9A1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1
|
|
chr7_-_138665853
|
0.177
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr2_+_26568900
|
0.177
|
NM_033505
|
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chr17_+_72244504
|
0.176
|
NM_052869
|
TTYH2
|
tweety homolog 2 (Drosophila)
|
|
chr6_+_19837599
|
0.176
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr10_-_113943467
|
0.174
|
NM_001244949
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr5_-_49737216
|
0.174
|
NM_198449
|
EMB
|
embigin
|
|
chr15_+_94841429
|
0.172
|
NM_001159643
NM_018349
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr13_+_73633097
|
0.171
|
NM_001730
|
KLF5
|
Kruppel-like factor 5 (intestinal)
|
|
chr9_+_131843378
|
0.170
|
NM_001135917
NM_020438
|
DOLPP1
|
dolichyl pyrophosphate phosphatase 1
|
|
chr1_-_19282773
|
0.168
|
NM_001136265
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr17_-_6459746
|
0.168
|
NM_001165966
NM_031220
|
PITPNM3
|
PITPNM family member 3
|
|
chr1_-_48462561
|
0.165
|
NM_001194986
|
LOC388630
|
UPF0632 protein A
|
|
chr16_-_17564737
|
0.163
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr10_+_24497719
|
0.161
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr14_-_78083109
|
0.161
|
NM_004863
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2
|
|
chr13_-_76055872
|
0.159
|
NM_014832
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr1_-_109935869
|
0.159
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chr2_+_121103670
|
0.158
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr1_-_109940497
|
0.158
|
NM_002959
|
SORT1
|
sortilin 1
|
|
chr13_+_113344354
|
0.156
|
NM_015205
NM_032189
|
ATP11A
|
ATPase, class VI, type 11A
|
|
chr17_+_12569206
|
0.155
|
NM_001146312
NM_153604
|
MYOCD
|
myocardin
|
|
chr1_+_62208091
|
0.155
|
NM_176877
|
INADL
|
InaD-like (Drosophila)
|
|
chr6_-_107435635
|
0.154
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr3_-_129612371
|
0.154
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr13_-_41240607
|
0.154
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr5_+_10353802
|
0.154
|
NM_005885
|
MARCH6
|
membrane-associated ring finger (C3HC4) 6
|
|
chr5_+_149109814
|
0.153
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr14_+_70078309
|
0.153
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr3_-_87040246
|
0.152
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr9_+_131799212
|
0.150
|
NM_032809
|
FAM73B
|
family with sequence similarity 73, member B
|
|
chr1_+_65613211
|
0.147
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr6_-_42419782
|
0.146
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr3_+_57994087
|
0.146
|
NM_001164317
NM_001164318
NM_001164319
NM_001457
|
FLNB
|
filamin B, beta
|
|
chr12_+_56521953
|
0.143
|
NM_001184796
NM_015292
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr5_+_135364583
|
0.143
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr22_+_21771661
|
0.141
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr1_+_65613771
|
0.141
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr1_+_37940074
|
0.140
|
NM_025079
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr5_-_141257943
|
0.140
|
NM_002587
NM_032420
|
PCDH1
|
protocadherin 1
|
|
chr3_+_43328001
|
0.140
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr2_+_46524448
|
0.140
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr13_+_96329392
|
0.139
|
NM_006260
|
DNAJC3
|
DnaJ (Hsp40) homolog, subfamily C, member 3
|
|
chr12_-_89746244
|
0.138
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr17_+_72209500
|
0.137
|
NM_032646
|
TTYH2
|
tweety homolog 2 (Drosophila)
|
|
chr15_+_73344824
|
0.136
|
NM_001172623
NM_001172624
NM_002499
|
NEO1
|
neogenin 1
|
|
chr7_-_139876718
|
0.135
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr6_+_80340971
|
0.134
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr11_-_87908551
|
0.134
|
NM_022337
|
RAB38
|
RAB38, member RAS oncogene family
|
|
chr20_-_6104056
|
0.133
|
NM_017671
|
FERMT1
|
fermitin family member 1
|
|
chr4_+_108745718
|
0.132
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr5_+_175875355
|
0.131
|
NM_014613
|
FAF2
|
Fas associated factor family member 2
|
|
chr10_+_119000583
|
0.131
|
NM_003054
|
SLC18A2
|
solute carrier family 18 (vesicular monoamine), member 2
|
|
chr2_-_38604430
|
0.130
|
NM_001135673
NM_022374
|
ATL2
|
atlastin GTPase 2
|
|
chr5_+_149151502
|
0.130
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr4_+_141445311
|
0.129
|
NM_153702
|
ELMOD2
|
ELMO/CED-12 domain containing 2
|
|
chr11_+_44785975
|
0.128
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr14_-_95786095
|
0.127
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr3_-_129599220
|
0.127
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr1_+_169075869
|
0.126
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr9_-_102861305
|
0.125
|
NM_015051
|
ERP44
|
endoplasmic reticulum protein 44
|
|
chr10_+_101419169
|
0.125
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr6_-_80656870
|
0.125
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr22_+_47022657
|
0.125
|
NM_015124
|
GRAMD4
|
GRAM domain containing 4
|
|
chr3_+_111578026
|
0.125
|
NM_001134438
NM_145753
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr7_+_23636970
|
0.124
|
NM_138771
|
CCDC126
|
coiled-coil domain containing 126
|
|
chr8_-_103424897
|
0.123
|
NM_015902
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5
|
|
chr1_+_116185249
|
0.123
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr20_-_41818464
|
0.122
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr1_-_6321034
|
0.122
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr16_+_71392615
|
0.122
|
NM_001740
NM_007088
|
CALB2
|
calbindin 2
|
|
chr2_+_191002485
|
0.121
|
NM_001042521
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr1_-_169455099
|
0.121
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr11_+_66886735
|
0.121
|
NM_012308
|
KDM2A
|
lysine (K)-specific demethylase 2A
|
|
chr13_-_30169795
|
0.121
|
NM_003045
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1
|
|
chr17_+_1959512
|
0.120
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr22_+_38302085
|
0.120
|
NM_033386
|
MICALL1
|
MICAL-like 1
|
|
chr2_+_48541767
|
0.120
|
NM_002158
|
FOXN2
|
forkhead box N2
|
|
chr2_+_109335906
|
0.119
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr2_+_7057471
|
0.119
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chrX_+_68725077
|
0.118
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr15_+_69591272
|
0.116
|
NM_017705
|
PAQR5
|
progestin and adipoQ receptor family member V
|
|
chr1_-_184943673
|
0.116
|
NM_052966
|
FAM129A
|
family with sequence similarity 129, member A
|
|
chr12_+_38710556
|
0.115
|
NM_001013620
|
ALG10B
|
asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog B (yeast)
|
|
chr11_+_60681370
|
0.113
|
NM_024092
|
TMEM109
|
transmembrane protein 109
|
|
chr2_-_1748285
|
0.111
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr1_+_107682628
|
0.111
|
NM_001113226
NM_001113228
NM_014917
|
NTNG1
|
netrin G1
|
|
chr4_+_86699846
|
0.110
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_-_175161832
|
0.110
|
NM_001162895
NM_014656
NM_001162893
NM_001162894
|
KIAA0040
|
KIAA0040
|
|
chr1_+_116184573
|
0.110
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr2_+_203499819
|
0.110
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr18_+_23806385
|
0.110
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr2_+_191014971
|
0.110
|
NM_001042520
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr10_-_125851850
|
0.110
|
NM_014863
NM_015892
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr6_-_128841432
|
0.109
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr11_-_113345841
|
0.109
|
NM_000795
NM_016574
|
DRD2
|
dopamine receptor D2
|
|
chr19_+_41222915
|
0.108
|
NM_025194
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|
|
chr3_-_32544358
|
0.108
|
NM_017801
|
CMTM6
|
CKLF-like MARVEL transmembrane domain containing 6
|
|
chr9_-_34590137
|
0.108
|
NM_001207011
|
CNTFR
|
ciliary neurotrophic factor receptor
|
|
chr14_-_104313730
|
0.106
|
NM_015316
|
PPP1R13B
|
protein phosphatase 1, regulatory subunit 13B
|
|
chr9_-_34589679
|
0.106
|
NM_001842
NM_147164
|
CNTFR
|
ciliary neurotrophic factor receptor
|
|
chr4_+_86396251
|
0.105
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_-_108507474
|
0.105
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr3_-_107809754
|
0.103
|
NM_001777
NM_198793
|
CD47
|
CD47 molecule
|
|
chr7_-_158622202
|
0.103
|
NM_020728
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr9_+_131102786
|
0.102
|
NM_005094
|
SLC27A4
|
solute carrier family 27 (fatty acid transporter), member 4
|
|
chr14_-_95599794
|
0.101
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr10_+_71211215
|
0.101
|
NM_012339
|
TSPAN15
|
tetraspanin 15
|
|
chr11_+_94501503
|
0.101
|
NM_130847
|
AMOTL1
|
angiomotin like 1
|
|
chr2_+_191045588
|
0.101
|
NM_001042519
NM_032321
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr1_+_236849753
|
0.101
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr4_+_108814419
|
0.101
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr3_-_53080038
|
0.100
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr4_+_30721865
|
0.100
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr5_-_175964223
|
0.099
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr14_-_55369163
|
0.098
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr1_-_155948317
|
0.098
|
NM_001162383
NM_001162384
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|