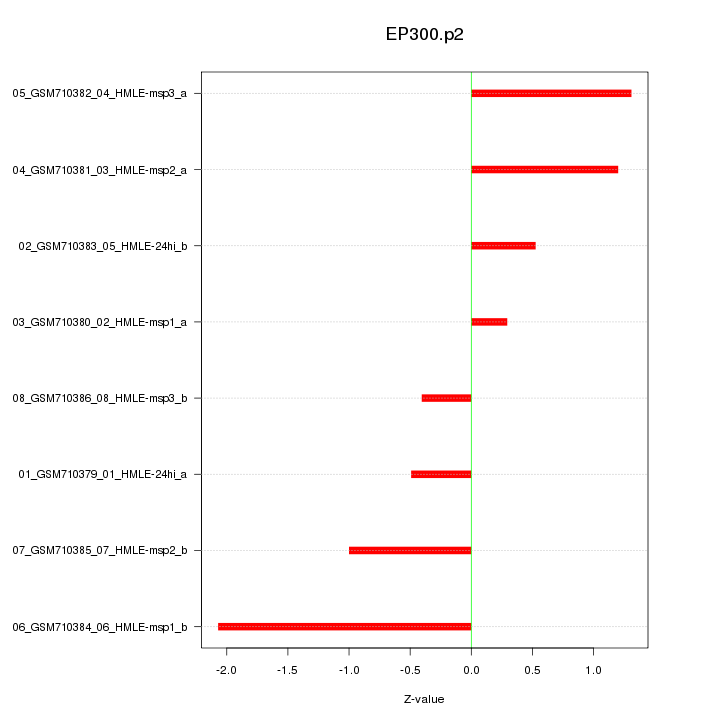
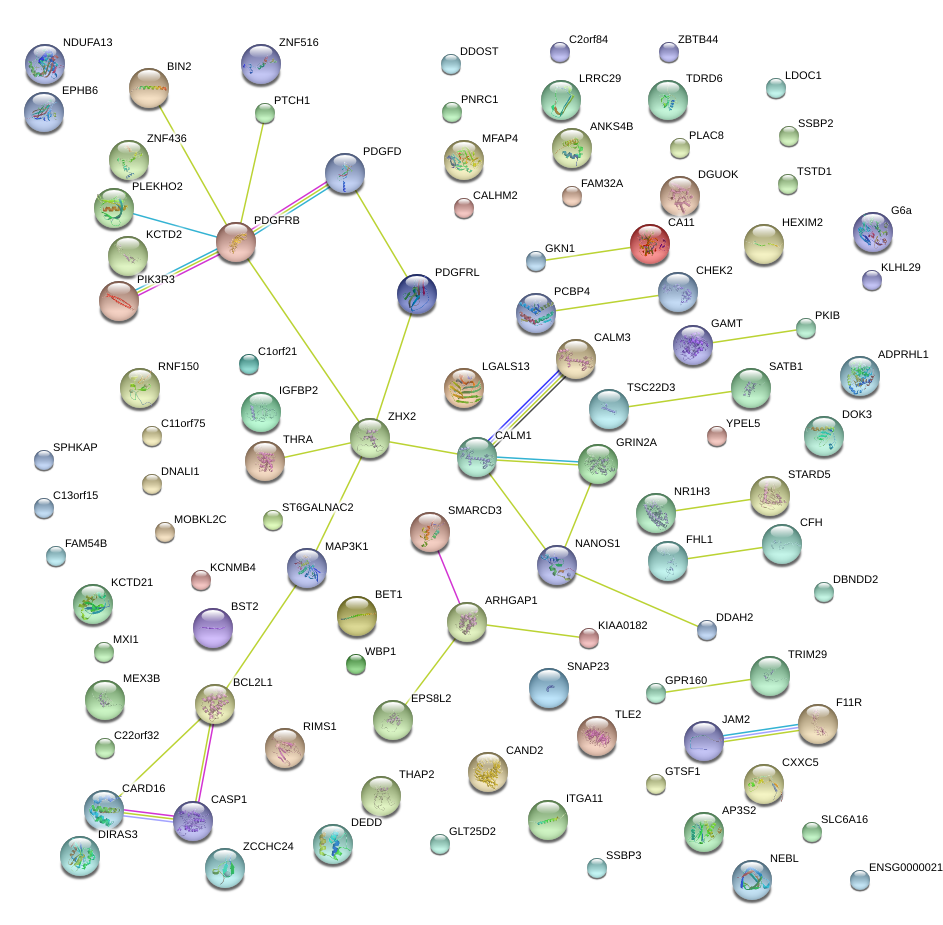
|
chr12_-_54867372
|
0.971
|
NM_144594
|
GTSF1
|
gametocyte specific factor 1
|
|
chr10_-_21462987
|
0.577
|
|
NEBL
|
nebulette
|
|
chr1_-_161008733
|
0.526
|
NM_001113205
NM_001113206
NM_001113207
|
TSTD1
F11R
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1
F11 receptor
|
|
chr6_+_89790412
|
0.401
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr10_-_21463019
|
0.377
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr19_+_16296211
|
0.367
|
NM_014077
|
FAM32A
|
family with sequence similarity 32, member A
|
|
chr6_+_89790464
|
0.366
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr10_+_111985628
|
0.354
|
NM_005962
|
MXI1
|
MAX interactor 1
|
|
chr5_-_149535309
|
0.320
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr6_+_122793061
|
0.308
|
NM_181794
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta
|
|
chrX_-_140271293
|
0.299
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr12_+_72057676
|
0.289
|
NM_031435
|
THAP2
|
THAP domain containing, apoptosis associated protein 2
|
|
chr6_-_31698033
|
0.277
|
NM_013974
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2
|
|
chr2_-_99224868
|
0.271
|
NM_001008215
|
COA5
|
cytochrome C oxidase assembly factor 5
|
|
chr2_-_27294491
|
0.269
|
NM_001134693
|
OST4
|
oligosaccharyltransferase 4 homolog (S. cerevisiae)
|
|
chr1_+_26146446
|
0.266
|
|
FAM54B
|
family with sequence similarity 54, member B
|
|
chr15_-_81616439
|
0.265
|
NM_181900
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5
|
|
chr11_-_93276513
|
0.263
|
NM_020179
|
C11orf75
|
chromosome 11 open reading frame 75
|
|
chr6_+_46655611
|
0.261
|
NM_001010870
NM_001168359
|
TDRD6
|
tudor domain containing 6
|
|
chr15_-_82338348
|
0.260
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr3_+_169755733
|
0.256
|
NM_014373
|
GPR160
|
G protein-coupled receptor 160
|
|
chr1_+_184356186
|
0.255
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr22_+_27053392
|
0.251
|
|
MIAT
|
myocardial infarction associated transcript (non-protein coding)
|
|
chr6_-_31697953
|
0.246
|
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2
|
|
chr3_+_169755782
|
0.246
|
|
GPR160
|
G protein-coupled receptor 160
|
|
chr15_-_82338459
|
0.244
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr16_-_67260720
|
0.240
|
NM_012163
|
LRRC29
|
leucine rich repeat containing 29
|
|
chr5_-_149535138
|
0.236
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr1_+_26146622
|
0.235
|
NM_019557
|
FAM54B
|
family with sequence similarity 54, member B
|
|
chr19_-_49828434
|
0.234
|
NM_014037
|
SLC6A16
|
solute carrier family 6, member 16
|
|
chr2_+_217498082
|
0.229
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chrX_-_140271275
|
0.227
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chrX_+_135229536
|
0.222
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr6_+_72596405
|
0.218
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_-_47082529
|
0.218
|
NM_201403
|
MOB3C
|
MOB kinase activator 3C
|
|
chr5_+_56111379
|
0.212
|
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr19_-_1401474
|
0.209
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr1_-_23694497
|
0.205
|
NM_030634
|
ZNF436
|
zinc finger protein 436
|
|
chr9_-_98270321
|
0.203
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr19_+_19627018
|
0.201
|
NM_015965
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13
|
|
chr1_+_184356227
|
0.200
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr16_-_10275743
|
0.200
|
NM_001134408
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr12_+_70759974
|
0.199
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr11_+_47269850
|
0.198
|
NM_001251934
NM_001251935
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr11_-_104035023
|
0.198
|
NM_025208
NM_033135
|
PDGFD
|
platelet derived growth factor D
|
|
chr11_-_104905849
|
0.196
|
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr1_+_26146376
|
0.195
|
NM_001099625
NM_001099627
|
FAM54B
|
family with sequence similarity 54, member B
|
|
chr8_+_123794043
|
0.194
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr4_-_84030995
|
0.192
|
NM_001130716
|
PLAC8
|
placenta-specific 8
|
|
chr2_+_74153987
|
0.191
|
|
DGUOK
|
deoxyguanosine kinase
|
|
chr7_+_142552547
|
0.191
|
NM_004445
|
EPHB6
|
EPH receptor B6
|
|
chr15_+_65134081
|
0.190
|
NM_001195059
NM_025201
|
PLEKHO2
|
pleckstrin homology domain containing, family O member 2
|
|
chr10_-_81205091
|
0.190
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr11_-_104034685
|
0.188
|
|
PDGFD
|
platelet derived growth factor D
|
|
chr5_-_81046943
|
0.185
|
NM_012446
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr11_+_706607
|
0.183
|
|
EPS8L2
|
EPS8-like 2
|
|
chr2_-_25016214
|
0.183
|
NM_001013663
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1
|
|
chr2_+_74154015
|
0.182
|
|
DGUOK
|
deoxyguanosine kinase
|
|
chr2_+_74685526
|
0.180
|
NM_012477
|
WBP1
|
WW domain binding protein 1
|
|
chr6_-_31697568
|
0.179
|
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2
|
|
chr1_-_184006491
|
0.177
|
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr15_+_42787850
|
0.177
|
|
SNAP23
|
synaptosomal-associated protein, 23kDa
|
|
chr10_-_105212149
|
0.176
|
NM_015916
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr10_+_120789227
|
0.175
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr2_+_74153931
|
0.175
|
NM_080916
NM_080918
|
DGUOK
|
deoxyguanosine kinase
|
|
chr3_-_51996855
|
0.175
|
NM_033010
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr12_-_51717941
|
0.174
|
|
BIN2
|
bridging integrator 2
|
|
chr16_+_85646923
|
0.174
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr1_-_46598250
|
0.173
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr7_-_150974179
|
0.173
|
NM_003078
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr1_-_68516311
|
0.173
|
NM_004675
|
DIRAS3
|
DIRAS family, GTP-binding RAS-like 3
|
|
chr11_-_77899640
|
0.172
|
NM_001029859
|
KCTD21
|
potassium channel tetramerisation domain containing 21
|
|
chr7_-_93633527
|
0.172
|
NM_005868
|
BET1
|
blocked early in transport 1 homolog (S. cerevisiae)
|
|
chr11_-_130184314
|
0.171
|
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr2_+_30369868
|
0.171
|
|
YPEL5
|
yippee-like 5 (Drosophila)
|
|
chr13_-_114103452
|
0.168
|
NM_199162
|
ADPRHL1
|
ADP-ribosylhydrolase like 1
|
|
chr11_-_2925174
|
0.168
|
NM_007105
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense
|
|
chr8_+_17434700
|
0.167
|
|
PDGFRL
|
platelet-derived growth factor receptor-like
|
|
chr17_-_19290471
|
0.167
|
NM_001198695
NM_002404
|
MFAP4
|
microfibrillar-associated protein 4
|
|
chr16_-_2336874
|
0.167
|
|
|
|
|
chr11_-_46722160
|
0.166
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr13_+_42031525
|
0.164
|
NM_014059
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr17_+_78388979
|
0.163
|
|
ENDOV
|
endonuclease V
|
|
chr13_+_42031679
|
0.162
|
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr19_-_3047632
|
0.162
|
NM_001144761
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr1_-_161008710
|
0.161
|
|
TSTD1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1
|
|
chr3_-_151987332
|
0.161
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr20_+_44034632
|
0.161
|
NM_001197139
NM_001048221
NM_001048222
NM_001197140
NM_018478
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr17_+_43238263
|
0.161
|
NM_144608
|
HEXIM2
|
hexamethylene bis-acetamide inducible 2
|
|
chr21_+_27011588
|
0.160
|
NM_021219
|
JAM2
|
junctional adhesion molecule 2
|
|
chr11_-_46722139
|
0.159
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr4_-_142054431
|
0.159
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr15_-_68724434
|
0.158
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr2_+_23608297
|
0.158
|
NM_052920
|
KLHL29
|
kelch-like 29 (Drosophila)
|
|
chr13_+_42031690
|
0.155
|
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr1_+_38022514
|
0.154
|
NM_003462
|
DNALI1
|
dynein, axonemal, light intermediate chain 1
|
|
chr11_-_120008738
|
0.152
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr5_+_139028483
|
0.152
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr20_-_30311693
|
0.151
|
|
BCL2L1
|
BCL2-like 1
|
|
chr22_-_29137776
|
0.150
|
NM_001005735
NM_007194
NM_145862
|
CHEK2
|
checkpoint kinase 2
|
|
chrX_-_107019001
|
0.148
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr3_-_18480203
|
0.147
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr5_-_176936857
|
0.146
|
NM_024872
|
DOK3
|
docking protein 3
|
|
chr19_+_47104389
|
0.146
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr2_+_24346339
|
0.146
|
NM_001145710
|
LOC375190
|
UPF0638 protein B
|
|
chr1_-_54871681
|
0.146
|
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr17_-_74582144
|
0.145
|
NM_006456
|
ST6GALNAC2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2
|
|
chr3_+_12838170
|
0.145
|
NM_001162499
NM_012298
|
CAND2
|
cullin-associated and neddylation-dissociated 2 (putative)
|
|
chr17_+_73043319
|
0.144
|
|
KCTD2
|
potassium channel tetramerisation domain containing 2
|
|
chr17_+_38230678
|
0.143
|
|
THRA
|
thyroid hormone receptor, alpha
|
|
chr19_+_18530423
|
0.143
|
|
|
|
|
chr19_-_49149252
|
0.143
|
NM_001217
|
CA11
|
carbonic anhydrase XI
|
|
chr5_-_176936834
|
0.143
|
|
DOK3
|
docking protein 3
|
|
chrX_+_135251454
|
0.142
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr17_+_78388944
|
0.142
|
NM_001164637
NM_001164638
NM_173627
|
ENDOV
|
endonuclease V
|
|
chr15_-_90437171
|
0.142
|
|
AP3S2
|
adaptor-related protein complex 3, sigma 2 subunit
|
|
chr18_-_74207145
|
0.141
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr19_-_17516383
|
0.141
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr2_-_229046313
|
0.140
|
NM_001142644
NM_030623
|
SPHKAP
|
SPHK1 interactor, AKAP domain containing
|
|
chr11_-_120008219
|
0.140
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr21_+_27011766
|
0.140
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr22_+_42475732
|
0.138
|
|
C22orf32
|
chromosome 22 open reading frame 32
|
|
chr1_-_161102367
|
0.137
|
|
DEDD
|
death effector domain containing
|
|
chr11_-_119999516
|
0.137
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr12_-_48744553
|
0.136
|
NM_001172681
NM_152320
|
ZNF641
|
zinc finger protein 641
|
|
chr19_+_9473695
|
0.136
|
NM_003451
|
ZNF177
|
zinc finger protein 177
|
|
chr9_+_130186652
|
0.135
|
NM_007135
|
ZNF79
|
zinc finger protein 79
|
|
chr2_+_233415300
|
0.135
|
NM_004846
|
EIF4E2
|
eukaryotic translation initiation factor 4E family member 2
|
|
chrX_-_107018889
|
0.135
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr3_-_178789535
|
0.134
|
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr20_+_44035203
|
0.133
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr2_+_71295685
|
0.133
|
|
NAGK
|
N-acetylglucosamine kinase
|
|
chr18_+_66382490
|
0.132
|
NM_001093729
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr16_-_31085503
|
0.132
|
NM_024706
|
ZNF668
|
zinc finger protein 668
|
|
chr21_-_38639625
|
0.132
|
NM_006052
|
DSCR3
|
Down syndrome critical region gene 3
|
|
chr15_+_74218786
|
0.131
|
NM_005576
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr21_-_45759234
|
0.130
|
|
C21orf2
|
chromosome 21 open reading frame 2
|
|
chr6_-_111136330
|
0.130
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr8_+_123793894
|
0.130
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr2_+_39892637
|
0.129
|
NM_001167959
|
TMEM178
|
transmembrane protein 178
|
|
chr10_-_45496259
|
0.128
|
NM_001039380
|
C10orf25
|
chromosome 10 open reading frame 25
|
|
chr11_-_120008801
|
0.127
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr15_+_74218794
|
0.127
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr12_+_7942025
|
0.127
|
|
NANOG
|
Nanog homeobox
|
|
chr21_-_38639593
|
0.127
|
|
DSCR3
|
Down syndrome critical region gene 3
|
|
chr1_+_6086072
|
0.127
|
NM_003636
NM_172130
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr11_+_64001974
|
0.127
|
NM_001243733
NM_003377
|
VEGFB
|
vascular endothelial growth factor B
|
|
chr2_+_217498205
|
0.126
|
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr4_-_156874993
|
0.126
|
NM_001334
|
CTSO
|
cathepsin O
|
|
chr22_+_42475661
|
0.126
|
NM_033318
|
C22orf32
|
chromosome 22 open reading frame 32
|
|
chr20_+_34680569
|
0.126
|
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr5_+_119799898
|
0.125
|
|
PRR16
|
proline rich 16
|
|
chr9_-_130667571
|
0.124
|
|
LOC100131355
ST6GALNAC6
|
uncharacterized LOC100131355
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr1_-_161102457
|
0.124
|
NM_001039711
NM_032998
|
DEDD
|
death effector domain containing
|
|
chr19_-_54984341
|
0.124
|
NM_145057
|
CDC42EP5
|
CDC42 effector protein (Rho GTPase binding) 5
|
|
chr11_-_34535302
|
0.123
|
NM_001243080
NM_001422
|
ELF5
|
E74-like factor 5 (ets domain transcription factor)
|
|
chr19_-_1605462
|
0.123
|
|
UQCR11
|
ubiquinol-cytochrome c reductase, complex III subunit XI
|
|
chr6_-_40555111
|
0.122
|
NM_020737
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2
|
|
chr8_-_91657898
|
0.122
|
NM_001008495
NM_001146273
|
TMEM64
|
transmembrane protein 64
|
|
chr10_-_64578926
|
0.122
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr11_-_67205151
|
0.121
|
NM_005608
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein
|
|
chr19_+_11039419
|
0.121
|
NM_138358
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr9_-_115774471
|
0.120
|
NM_001101338
|
ZNF883
|
zinc finger protein 883
|
|
chr8_+_145159304
|
0.120
|
NM_032272
|
MAF1
|
MAF1 homolog (S. cerevisiae)
|
|
chr1_+_236558715
|
0.119
|
NM_080738
|
EDARADD
|
EDAR-associated death domain
|
|
chrX_+_70315637
|
0.119
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr10_-_127511795
|
0.119
|
|
UROS
|
uroporphyrinogen III synthase
|
|
chr17_-_74581881
|
0.119
|
|
ST6GALNAC2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2
|
|
chr15_+_74218811
|
0.119
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr7_+_128864740
|
0.119
|
NM_001130720
NM_015328
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr22_+_24577417
|
0.119
|
|
SUSD2
|
sushi domain containing 2
|
|
chr10_+_81892497
|
0.119
|
|
PLAC9
|
placenta-specific 9
|
|
chr8_+_356807
|
0.119
|
NM_012173
NM_183420
NM_183421
|
FBXO25
|
F-box protein 25
|
|
chr1_+_221052735
|
0.118
|
NM_021958
|
HLX
|
H2.0-like homeobox
|
|
chr5_-_149535420
|
0.118
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr1_+_3541587
|
0.118
|
|
TPRG1L
|
tumor protein p63 regulated 1-like
|
|
chr17_+_7342688
|
0.118
|
NM_004112
|
FGF11
|
fibroblast growth factor 11
|
|
chr20_+_62688438
|
0.118
|
NM_198723
|
TCEA2
|
transcription elongation factor A (SII), 2
|
|
chr11_+_67250595
|
0.118
|
|
AIP
|
aryl hydrocarbon receptor interacting protein
|
|
chr15_-_93616358
|
0.118
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr7_-_45128492
|
0.118
|
NM_001146334
|
NACAD
|
NAC alpha domain containing
|
|
chr12_-_63328663
|
0.115
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr12_-_92539614
|
0.115
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr22_-_19435211
|
0.115
|
|
C22orf39
HIRA
|
chromosome 22 open reading frame 39
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr1_+_111415721
|
0.114
|
NM_000560
|
CD53
|
CD53 molecule
|
|
chr11_-_63933493
|
0.114
|
NM_014067
|
MACROD1
|
MACRO domain containing 1
|
|
chr11_+_134094565
|
0.114
|
|
VPS26B
|
vacuolar protein sorting 26 homolog B (S. pombe)
|
|
chr15_-_93616308
|
0.114
|
|
RGMA
|
RGM domain family, member A
|
|
chr3_+_134514300
|
0.113
|
|
EPHB1
|
EPH receptor B1
|
|
chr1_-_173886401
|
0.113
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr9_+_130186714
|
0.113
|
|
ZNF79
|
zinc finger protein 79
|
|
chr11_+_2924504
|
0.113
|
|
SLC22A18
|
solute carrier family 22, member 18
|
|
chr15_+_74287132
|
0.112
|
|
PML
|
promyelocytic leukemia
|
|
chr16_-_66952729
|
0.112
|
NM_001204744
NM_001204745
NM_001204746
NM_004062
|
CDH16
|
cadherin 16, KSP-cadherin
|
|
chr8_+_145159398
|
0.112
|
|
MAF1
|
MAF1 homolog (S. cerevisiae)
|
|
chr7_+_130126035
|
0.111
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr5_+_119800004
|
0.111
|
NM_016644
|
PRR16
|
proline rich 16
|
|
chr1_-_161102210
|
0.111
|
NM_001039712
|
DEDD
|
death effector domain containing
|
|
chr3_+_52279742
|
0.111
|
NM_144641
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M
|
|
chr14_-_68141205
|
0.110
|
|
VTI1B
|
vesicle transport through interaction with t-SNAREs homolog 1B (yeast)
|