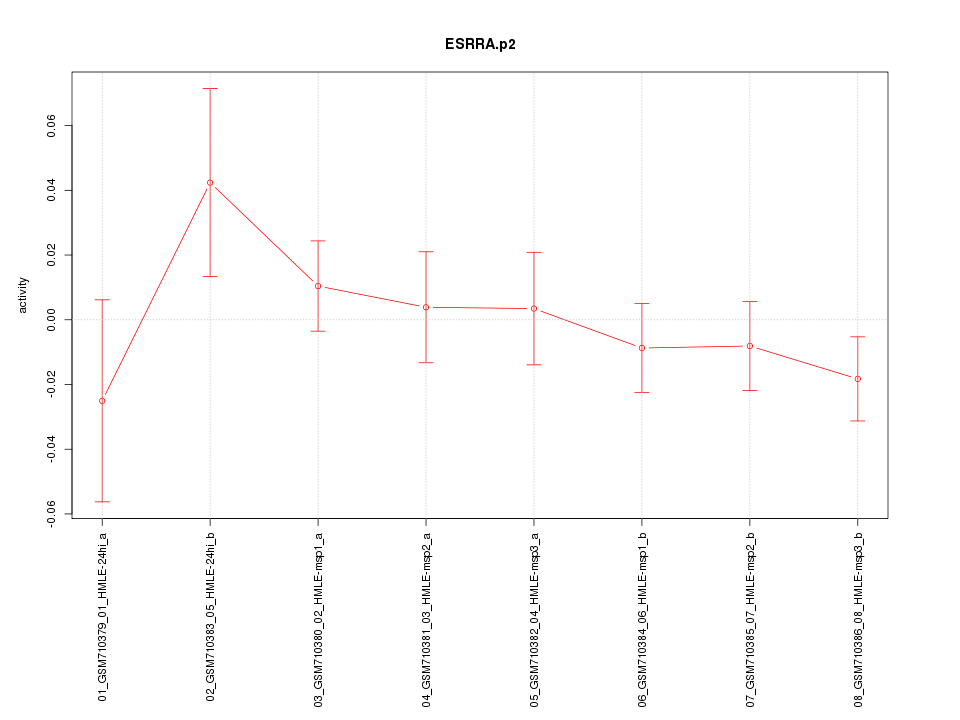
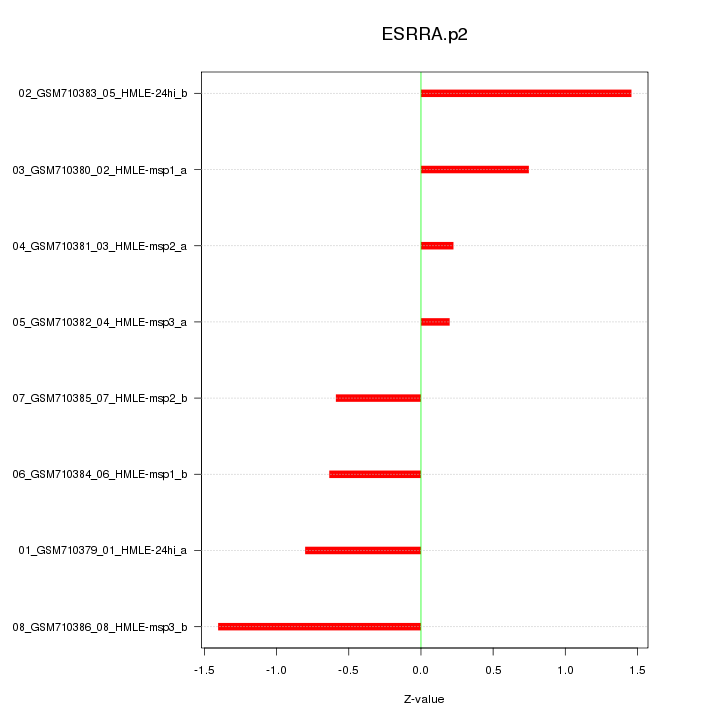
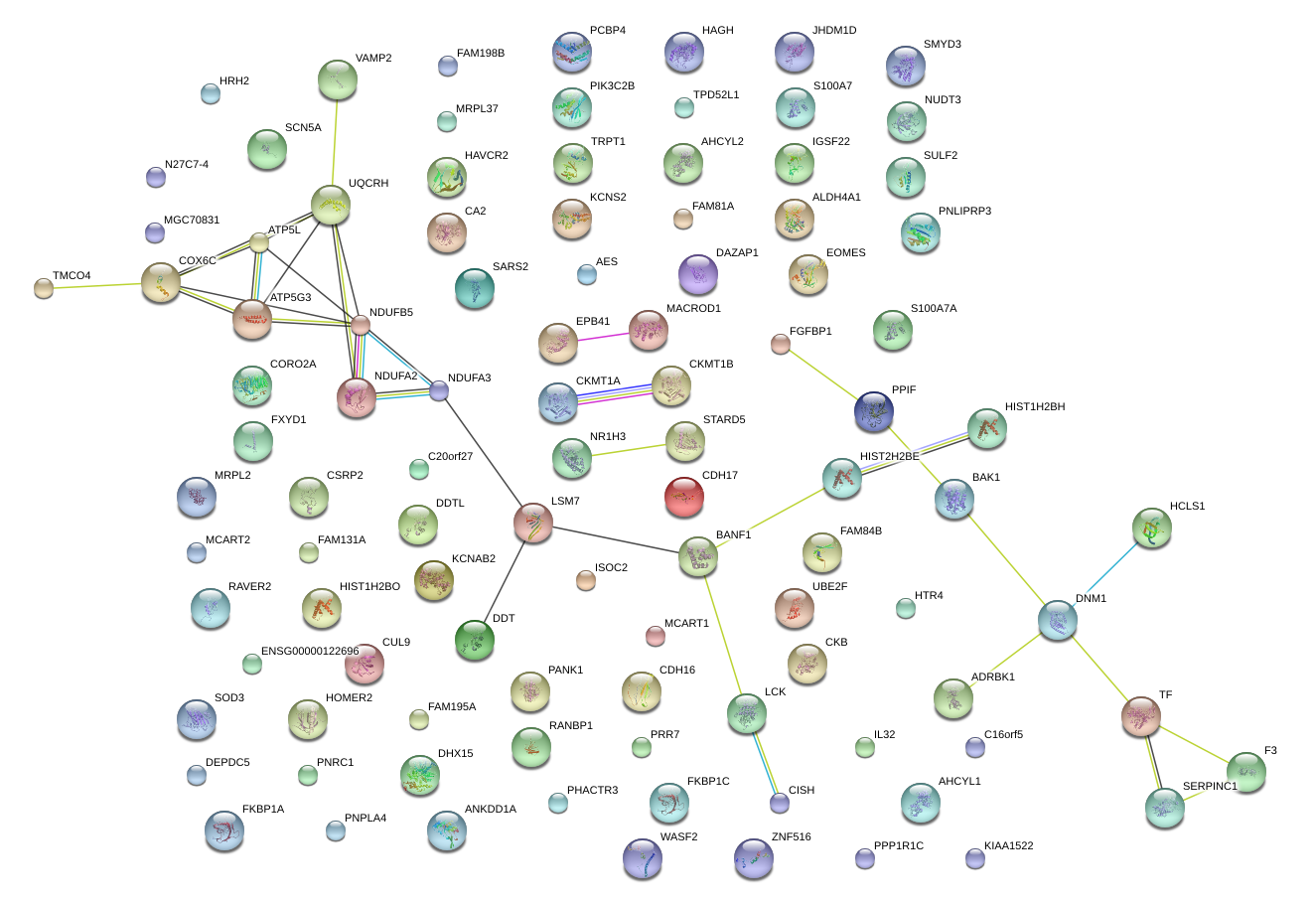
|
chr19_+_35629727
|
0.747
|
NM_021902
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr20_+_58179601
|
0.721
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr15_-_81616439
|
0.636
|
NM_181900
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5
|
|
chr15_-_83621350
|
0.596
|
NM_004839
NM_199330
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr15_+_43885251
|
0.555
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr16_-_4588379
|
0.504
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr15_+_43985083
|
0.478
|
NM_001015001
|
CKMT1A
|
creatine kinase, mitochondrial 1A
|
|
chr3_-_121379701
|
0.469
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr1_+_29213619
|
0.466
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr6_-_34360310
|
0.441
|
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3
|
|
chr15_+_59730272
|
0.422
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr14_-_103989112
|
0.410
|
|
CKB
|
creatine kinase, brain
|
|
chr6_-_43027114
|
0.406
|
NM_015950
|
MRPL2
|
mitochondrial ribosomal protein L2
|
|
chr1_-_173886401
|
0.390
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr1_+_29213569
|
0.388
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr11_+_47270433
|
0.385
|
NM_001130102
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr6_+_89790412
|
0.383
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr16_+_691827
|
0.383
|
NM_138418
|
FAM195A
|
family with sequence similarity 195, member A
|
|
chr11_-_63933204
|
0.378
|
|
MACROD1
|
MACRO domain containing 1
|
|
chr8_-_127570710
|
0.373
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr6_-_34360098
|
0.366
|
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3
|
|
chr6_+_89790464
|
0.357
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr10_+_118187381
|
0.354
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr8_-_100905910
|
0.347
|
|
COX6C
|
cytochrome c oxidase subunit VIc
|
|
chr16_+_691901
|
0.345
|
|
FAM195A
|
family with sequence similarity 195, member A
|
|
chr1_+_6094342
|
0.340
|
NM_001199860
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr1_-_20126368
|
0.331
|
NM_181719
|
TMCO4
|
transmembrane and coiled-coil domains 4
|
|
chr16_+_3115312
|
0.323
|
NM_001012631
NM_001012633
NM_001012718
NM_004221
|
IL32
|
interleukin 32
|
|
chr7_+_129008044
|
0.322
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr9_-_100954883
|
0.314
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr3_+_179322572
|
0.314
|
NM_001199957
NM_001199958
NM_002492
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa
|
|
chr18_-_74207145
|
0.313
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr8_-_127570465
|
0.312
|
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr5_-_148034089
|
0.311
|
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr1_+_65210724
|
0.308
|
NM_018211
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr2_-_176046120
|
0.302
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr14_-_103989195
|
0.299
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chr9_+_130965666
|
0.298
|
|
DNM1
|
dynamin 1
|
|
chr8_+_86376130
|
0.293
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr20_-_46415277
|
0.287
|
NM_001161841
NM_198596
|
SULF2
|
sulfatase 2
|
|
chr20_-_1373571
|
0.286
|
|
FKBP1A
|
FK506 binding protein 1A, 12kDa
|
|
chr19_-_55973048
|
0.284
|
NM_001136201
NM_001136202
NM_024710
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr19_-_3062191
|
0.275
|
NM_001130
NM_198970
|
AES
|
amino-terminal enhancer of split
|
|
chr5_+_176875325
|
0.273
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr1_+_33231260
|
0.272
|
|
KIAA1522
|
KIAA1522
|
|
chr3_-_121379757
|
0.271
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr1_-_19217384
|
0.270
|
NM_001161504
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr3_-_27763784
|
0.268
|
NM_005442
|
EOMES
|
eomesodermin
|
|
chr16_-_66952729
|
0.267
|
NM_001204744
NM_001204745
NM_001204746
NM_004062
|
CDH16
|
cadherin 16, KSP-cadherin
|
|
chr4_-_159094163
|
0.267
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr5_+_175084984
|
0.267
|
NM_001131055
|
HRH2
|
histamine receptor H2
|
|
chr1_+_33231227
|
0.262
|
NM_001198972
NM_001198973
|
KIAA1522
|
KIAA1522
|
|
chrX_-_7895764
|
0.262
|
NM_001142389
NM_001172672
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr9_-_37904331
|
0.259
|
NM_033412
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chr22_-_24110054
|
0.255
|
NM_213720
|
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10
|
|
chr3_-_38691118
|
0.252
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr3_-_50649080
|
0.252
|
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr22_-_24316602
|
0.249
|
NM_001084392
|
DDT
|
D-dopachrome tautomerase
|
|
chr11_+_67033889
|
0.249
|
NM_001619
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr22_+_20105076
|
0.249
|
|
RANBP1
|
RAN binding protein 1
|
|
chr19_+_1407750
|
0.248
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr11_-_63993340
|
0.248
|
NM_001160393
|
TRPT1
|
tRNA phosphotransferase 1
|
|
chr11_-_63993562
|
0.247
|
NM_001033678
NM_001160389
NM_001160390
NM_001160392
NM_031472
|
TRPT1
|
tRNA phosphotransferase 1
|
|
chr19_-_39443326
|
0.246
|
NM_148169
|
FBXO17
|
F-box protein 17
|
|
chr11_+_67033929
|
0.246
|
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr3_+_133465098
|
0.244
|
|
TF
|
transferrin
|
|
chr1_-_246580710
|
0.244
|
NM_022743
|
SMYD3
|
SET and MYND domain containing 3
|
|
chr6_+_43149925
|
0.242
|
|
CUL9
|
cullin 9
|
|
chr22_+_20105068
|
0.240
|
|
RANBP1
|
RAN binding protein 1
|
|
chr1_+_46769336
|
0.240
|
NM_006004
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein
|
|
chr20_-_3748355
|
0.236
|
NM_001039140
|
C20orf27
|
chromosome 20 open reading frame 27
|
|
chr11_+_118272103
|
0.236
|
NM_006476
|
ATP5L
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G
|
|
chr22_+_24309025
|
0.236
|
NM_001084393
|
DDTL
|
D-dopachrome tautomerase-like
|
|
chr19_-_55972997
|
0.236
|
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr1_+_32716839
|
0.231
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr10_+_81107242
|
0.224
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr3_-_51996855
|
0.222
|
NM_033010
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr19_-_55972935
|
0.220
|
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr19_-_3062911
|
0.219
|
NM_198969
|
AES
|
amino-terminal enhancer of split
|
|
chr22_+_20105145
|
0.219
|
|
RANBP1
|
RAN binding protein 1
|
|
chr6_+_125475334
|
0.219
|
NM_001003395
|
TPD52L1
|
tumor protein D52-like 1
|
|
chr22_+_32150002
|
0.218
|
NM_001007188
NM_001242896
NM_014662
|
DEPDC5
|
DEP domain containing 5
|
|
chr19_-_2328566
|
0.216
|
NM_016199
|
LSM7
|
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr16_-_1877140
|
0.216
|
NM_001040427
NM_005326
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr22_+_20105095
|
0.216
|
|
RANBP1
|
RAN binding protein 1
|
|
chr5_-_140027167
|
0.214
|
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa
|
|
chr22_+_32149962
|
0.213
|
|
DEPDC5
|
DEP domain containing 5
|
|
chr6_-_33547833
|
0.213
|
|
BAK1
|
BCL2-antagonist/killer 1
|
|
chr1_-_204459445
|
0.212
|
NM_002646
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr8_+_99439249
|
0.211
|
NM_020697
|
KCNS2
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2
|
|
chr11_+_65769956
|
0.210
|
|
BANF1
|
barrier to autointegration factor 1
|
|
chr15_+_65204053
|
0.209
|
NM_182703
|
ANKDD1A
|
ankyrin repeat and death domain containing 1A
|
|
chr19_+_54606141
|
0.209
|
NM_004542
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa
|
|
chr17_-_8066254
|
0.209
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr1_+_153388999
|
0.205
|
NM_176823
|
S100A7A
|
S100 calcium binding protein A7A
|
|
chr10_-_91404901
|
0.203
|
|
PANK1
|
pantothenate kinase 1
|
|
chr3_+_184055275
|
0.202
|
NM_144635
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr10_+_81107259
|
0.202
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr22_+_20104810
|
0.201
|
NM_002882
|
RANBP1
|
RAN binding protein 1
|
|
chr7_-_139876718
|
0.200
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr22_+_32149936
|
0.200
|
NM_001242897
|
DEPDC5
|
DEP domain containing 5
|
|
chr4_-_15939949
|
0.199
|
|
FGFBP1
|
fibroblast growth factor binding protein 1
|
|
chr5_-_156536185
|
0.199
|
NM_032782
|
HAVCR2
|
hepatitis A virus cellular receptor 2
|
|
chr1_-_27732069
|
0.198
|
|
WASF2
|
WAS protein family, member 2
|
|
chr12_-_77272767
|
0.198
|
NM_001321
|
CSRP2
|
cysteine and glycine-rich protein 2
|
|
chr2_+_238875689
|
0.197
|
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative)
|
|
chr8_-_100905865
|
0.197
|
|
COX6C
|
cytochrome c oxidase subunit VIc
|
|
chr11_-_18747718
|
0.196
|
NM_173588
|
IGSF22
|
immunoglobulin superfamily, member 22
|
|
chr2_+_182850550
|
0.195
|
NM_001080545
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C
|
|
chr4_+_24797203
|
0.194
|
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr6_+_26251826
|
0.194
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr9_-_130890007
|
0.194
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr9_-_130890026
|
0.194
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr15_-_74753490
|
0.194
|
NM_032907
NM_201265
|
UBL7
|
ubiquitin-like 7 (bone marrow stromal cell-derived)
|
|
chr9_-_130889965
|
0.193
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr4_+_24797084
|
0.192
|
NM_003102
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr2_+_219135114
|
0.192
|
NM_001077399
NM_015488
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr1_+_159557615
|
0.191
|
NM_001639
|
APCS
|
amyloid P component, serum
|
|
chr12_+_133067156
|
0.190
|
NM_001142641
|
FBRSL1
|
fibrosin-like 1
|
|
chr11_+_65769886
|
0.189
|
|
BANF1
|
barrier to autointegration factor 1
|
|
chr14_-_103988829
|
0.189
|
|
CKB
|
creatine kinase, brain
|
|
chr12_+_51632654
|
0.186
|
|
DAZAP2
|
DAZ associated protein 2
|
|
chr11_+_63993720
|
0.186
|
NM_001128612
NM_001128613
NM_032344
|
NUDT22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22
|
|
chr9_-_130679210
|
0.185
|
NM_175039
NM_175040
|
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4
|
|
chr17_+_79679370
|
0.185
|
NM_012140
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10
|
|
chr5_+_167718485
|
0.184
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr12_+_51632589
|
0.183
|
|
DAZAP2
|
DAZ associated protein 2
|
|
chr9_-_117150217
|
0.183
|
|
AKNA
|
AT-hook transcription factor
|
|
chr11_-_111957417
|
0.181
|
NM_012459
|
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B (yeast)
|
|
chr7_+_45614179
|
0.180
|
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr3_-_71778268
|
0.177
|
NM_001134650
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr9_-_130889993
|
0.177
|
|
PTGES2
|
prostaglandin E synthase 2
|
|
chr20_-_633844
|
0.176
|
NM_080725
|
SRXN1
|
sulfiredoxin 1
|
|
chr1_+_236558715
|
0.176
|
NM_080738
|
EDARADD
|
EDAR-associated death domain
|
|
chr5_+_85913762
|
0.175
|
NM_001867
|
COX7C
|
cytochrome c oxidase subunit VIIc
|
|
chr1_+_228270802
|
0.174
|
NM_001024227
NM_001024228
|
ARF1
|
ADP-ribosylation factor 1
|
|
chr8_+_120220554
|
0.174
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr1_+_16083132
|
0.174
|
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr4_-_140216951
|
0.174
|
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa
|
|
chr19_+_15052298
|
0.174
|
NM_012377
|
OR7C2
|
olfactory receptor, family 7, subfamily C, member 2
|
|
chrX_+_154255063
|
0.171
|
NM_023934
|
FUNDC2
|
FUN14 domain containing 2
|
|
chr1_-_205719340
|
0.171
|
NM_022731
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr2_-_74374998
|
0.170
|
NM_001035505
NM_212552
|
BOLA3
|
bolA homolog 3 (E. coli)
|
|
chr9_-_117156684
|
0.170
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr7_+_56032065
|
0.170
|
NM_001202469
NM_001483
|
GBAS
|
glioblastoma amplified sequence
|
|
chr9_-_4742042
|
0.169
|
NM_001199853
|
AK3
|
adenylate kinase 3
|
|
chr1_+_24829299
|
0.168
|
NM_001251977
NM_001251981
NM_001251983
NM_013441
|
RCAN3
|
RCAN family member 3
|
|
chr10_-_105156186
|
0.167
|
NM_001206426
NM_001206427
NM_032747
|
USMG5
|
up-regulated during skeletal muscle growth 5 homolog (mouse)
|
|
chr16_-_15149827
|
0.167
|
NM_173474
|
NTAN1
|
N-terminal asparagine amidase
|
|
chr1_-_173886464
|
0.167
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chrX_+_23801431
|
0.166
|
|
SAT1
|
spermidine/spermine N1-acetyltransferase 1
|
|
chr8_-_131028674
|
0.165
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr3_+_158288952
|
0.165
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chr19_-_55668956
|
0.165
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr15_+_78441737
|
0.165
|
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr20_-_633808
|
0.164
|
|
SRXN1
|
sulfiredoxin 1
|
|
chr8_+_124428964
|
0.164
|
NM_018024
|
WDYHV1
|
WDYHV motif containing 1
|
|
chr15_-_91475653
|
0.163
|
NM_198527
|
HDDC3
|
HD domain containing 3
|
|
chr19_+_1383898
|
0.163
|
|
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
|
|
chr22_+_44395090
|
0.163
|
NM_001003828
|
PARVB
|
parvin, beta
|
|
chr15_-_76352068
|
0.163
|
|
NRG4
|
neuregulin 4
|
|
chr3_+_159557649
|
0.162
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr17_+_13972718
|
0.162
|
NM_001303
|
COX10
|
COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast)
|
|
chr14_+_75898836
|
0.162
|
NM_001135049
|
JDP2
|
Jun dimerization protein 2
|
|
chr9_-_112191105
|
0.161
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chrX_-_15511358
|
0.160
|
|
PIR
|
pirin (iron-binding nuclear protein)
|
|
chr10_-_76995685
|
0.160
|
NM_144589
|
COMTD1
|
catechol-O-methyltransferase domain containing 1
|
|
chr6_+_43149905
|
0.159
|
NM_015089
|
CUL9
|
cullin 9
|
|
chr1_+_152881022
|
0.159
|
NM_005547
|
IVL
|
involucrin
|
|
chr6_-_34664587
|
0.159
|
NM_022758
NM_024294
|
C6orf106
|
chromosome 6 open reading frame 106
|
|
chr9_-_4741798
|
0.158
|
NM_001199855
|
AK3
|
adenylate kinase 3
|
|
chr16_+_21716283
|
0.158
|
NM_170664
|
OTOA
|
otoancorin
|
|
chr16_+_77756388
|
0.158
|
NM_001105663
NM_001243657
NM_001243660
NM_001243661
|
NUDT7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7
|
|
chr16_+_68679198
|
0.158
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr17_+_13972841
|
0.157
|
|
COX10
|
COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast)
|
|
chr6_-_34664517
|
0.156
|
|
C6orf106
|
chromosome 6 open reading frame 106
|
|
chrX_-_21776158
|
0.156
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr7_+_129007942
|
0.155
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr16_+_3115611
|
0.155
|
NM_001012635
|
IL32
|
interleukin 32
|
|
chr12_+_109915461
|
0.154
|
|
UBE3B
|
ubiquitin protein ligase E3B
|
|
chr19_+_1383904
|
0.154
|
|
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
|
|
chr16_+_776957
|
0.154
|
NM_032304
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr9_+_130965668
|
0.154
|
|
DNM1
|
dynamin 1
|
|
chr1_-_33502464
|
0.153
|
|
AK2
|
adenylate kinase 2
|
|
chr18_-_44497307
|
0.153
|
NM_004671
NM_173206
|
PIAS2
|
protein inhibitor of activated STAT, 2
|
|
chr14_+_42076763
|
0.152
|
NM_152447
|
LRFN5
|
leucine rich repeat and fibronectin type III domain containing 5
|
|
chrY_-_1460901
|
0.150
|
NM_001636
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6
|
|
chr2_-_74667553
|
0.149
|
NM_001015056
NM_033046
|
RTKN
|
rhotekin
|
|
chr15_-_66790068
|
0.149
|
NM_006049
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa
|
|
chr10_-_120938276
|
0.148
|
|
PRDX3
|
peroxiredoxin 3
|
|
chr15_-_65715385
|
0.148
|
NM_020962
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4
|
|
chr12_+_51632620
|
0.147
|
|
DAZAP2
|
DAZ associated protein 2
|
|
chr9_+_130965608
|
0.147
|
NM_001005336
NM_004408
|
DNM1
|
dynamin 1
|
|
chr19_+_1383877
|
0.147
|
NM_024407
|
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
|
|
chr6_+_43968303
|
0.146
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr2_-_172967627
|
0.146
|
|
DLX2
|
distal-less homeobox 2
|
|
chr19_+_2249112
|
0.145
|
NM_000479
|
AMH
|
anti-Mullerian hormone
|
|
chrX_+_106161589
|
0.145
|
NM_001171095
|
CLDN2
|
claudin 2
|
|
chr1_-_202927699
|
0.145
|
NM_001127687
|
ADIPOR1
|
adiponectin receptor 1
|
|
chrX_-_134185978
|
0.145
|
NM_001078172
NM_001134321
|
FAM127B
|
family with sequence similarity 127, member B
|