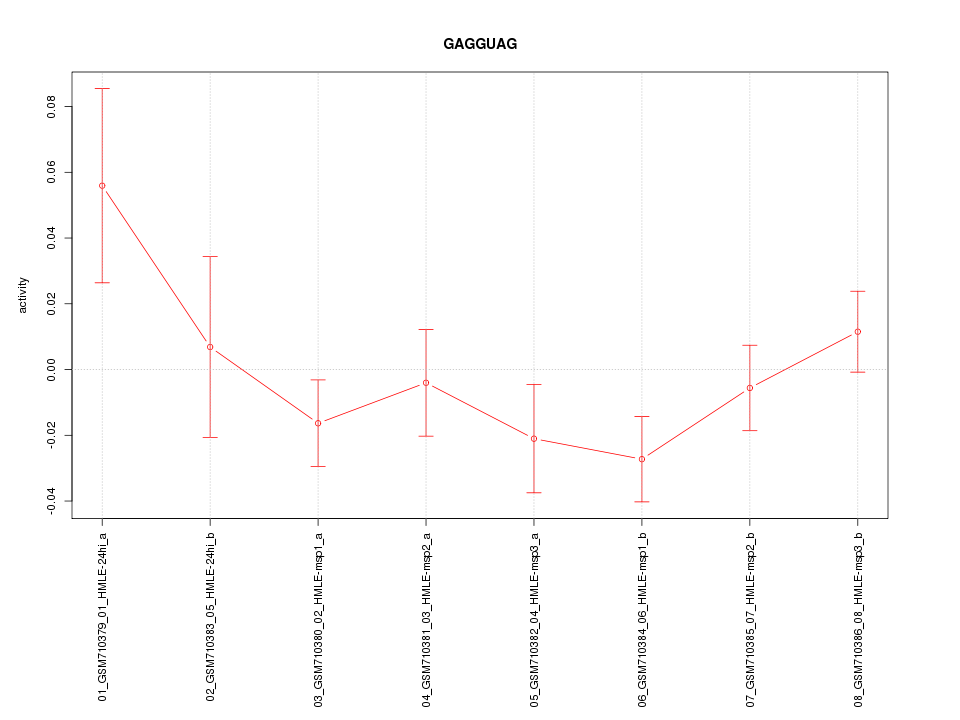
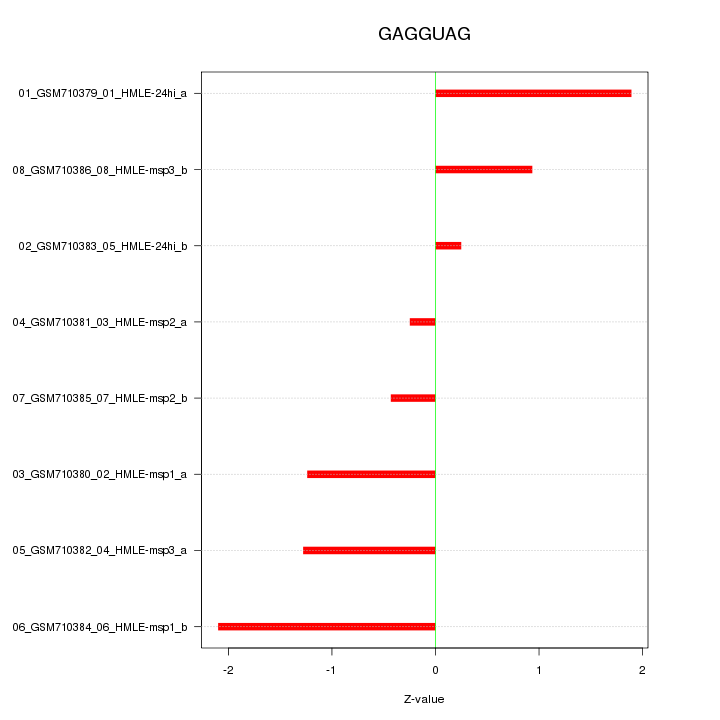
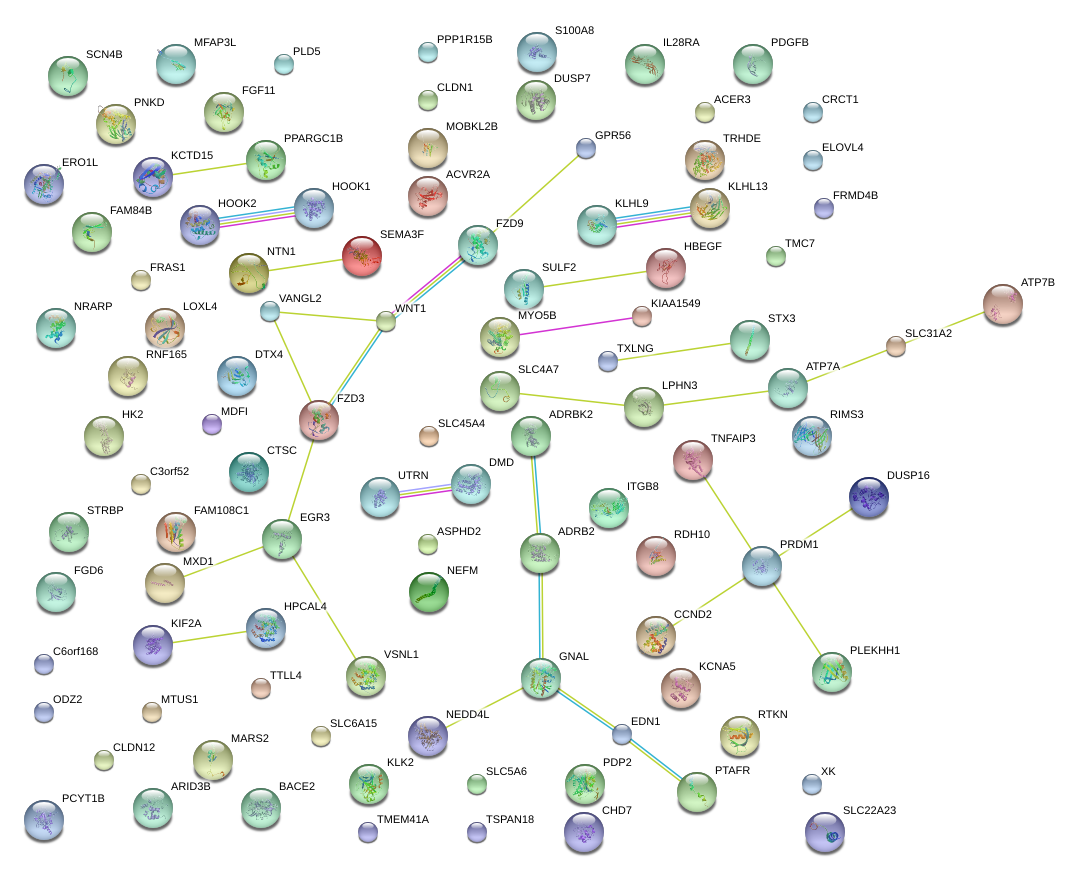
|
chr1_-_242687997
|
1.974
|
NM_152666
|
PLD5
|
phospholipase D family, member 5
|
|
chr1_-_242612757
|
1.820
|
NM_001195811
NM_001195812
|
PLD5
|
phospholipase D family, member 5
|
|
chr10_-_100027943
|
1.687
|
NM_032211
|
LOXL4
|
lysyl oxidase-like 4
|
|
chr9_-_27529434
|
1.549
|
NM_024761
|
MOB3B
|
MOB kinase activator 3B
|
|
chr1_+_60280462
|
1.240
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr3_-_69435429
|
1.157
|
NM_015123
|
FRMD4B
|
FERM domain containing 4B
|
|
chr8_+_28351694
|
1.156
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr1_-_153363544
|
1.137
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr20_-_46415277
|
1.115
|
NM_001161841
NM_198596
|
SULF2
|
sulfatase 2
|
|
chr20_-_46414636
|
1.110
|
NM_018837
|
SULF2
|
sulfatase 2
|
|
chr12_+_4382882
|
1.084
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr1_-_28503454
|
1.043
|
NM_000952
NM_001164723
|
PTAFR
|
platelet-activating factor receptor
|
|
chrX_+_37545008
|
1.014
|
NM_021083
|
XK
|
X-linked Kx blood group (McLeod syndrome)
|
|
chr5_+_166711842
|
0.999
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr2_+_17721806
|
0.998
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr1_+_152486977
|
0.980
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr18_+_11751470
|
0.949
|
NM_001142339
NM_002071
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr1_-_28520379
|
0.917
|
NM_001164721
NM_001164722
|
PTAFR
|
platelet-activating factor receptor
|
|
chr5_-_139726173
|
0.908
|
NM_001945
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr12_+_72666462
|
0.899
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr6_-_99797530
|
0.899
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr18_+_11689135
|
0.884
|
NM_182978
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr22_-_39636906
|
0.871
|
NM_033016
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr6_+_138188352
|
0.866
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr8_-_127570710
|
0.863
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr22_-_39640956
|
0.819
|
NM_002608
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr2_+_198570027
|
0.770
|
NM_138395
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial
|
|
chr14_+_67999915
|
0.763
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr3_+_111805174
|
0.760
|
NM_001171747
NM_024616
|
C3orf52
|
chromosome 3 open reading frame 52
|
|
chr5_+_149109814
|
0.750
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr9_+_115913227
|
0.744
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr8_+_24772454
|
0.734
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr8_-_142238672
|
0.734
|
NM_001080431
|
SLC45A4
|
solute carrier family 45, member 4
|
|
chr6_+_41606193
|
0.733
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr1_+_160370363
|
0.727
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr15_+_74833517
|
0.723
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr12_-_95611023
|
0.717
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr8_+_61591239
|
0.702
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr5_+_149151502
|
0.679
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr7_+_20370724
|
0.675
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr11_+_76571902
|
0.625
|
NM_018367
|
ACER3
|
alkaline ceramidase 3
|
|
chr1_-_175161832
|
0.623
|
NM_001162895
NM_014656
NM_001162893
NM_001162894
|
KIAA0040
|
KIAA0040
|
|
chr1_-_24513737
|
0.616
|
NM_170743
NM_173064
NM_173065
|
IL28RA
|
interleukin 28 receptor, alpha (interferon, lambda receptor)
|
|
chr18_-_47721165
|
0.604
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr8_+_24771268
|
0.600
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr1_-_41131323
|
0.588
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr12_-_85306579
|
0.586
|
NM_001146335
NM_018057
NM_182767
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chrX_-_24665454
|
0.576
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr12_+_49372235
|
0.570
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr17_+_8924822
|
0.569
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr11_-_88070905
|
0.569
|
NM_001114173
NM_001814
NM_148170
|
CTSC
|
cathepsin C
|
|
chr6_+_106534188
|
0.564
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr3_+_50192847
|
0.563
|
NM_004186
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr4_-_170947428
|
0.561
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr22_+_26825220
|
0.557
|
NM_020437
|
ASPHD2
|
aspartate beta-hydroxylase domain containing 2
|
|
chr8_+_74206772
|
0.551
|
NM_172037
|
RDH10
|
retinol dehydrogenase 10 (all-trans)
|
|
chrX_-_24690740
|
0.550
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr2_-_74667553
|
0.550
|
NM_001015056
NM_033046
|
RTKN
|
rhotekin
|
|
chr12_-_12715244
|
0.539
|
NM_030640
|
DUSP16
|
dual specificity phosphatase 16
|
|
chr8_-_22550708
|
0.537
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr21_+_42539727
|
0.537
|
NM_012105
NM_138991
NM_138992
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chrX_-_33357725
|
0.534
|
NM_000109
|
DMD
|
dystrophin
|
|
chr2_+_219575507
|
0.525
|
NM_014640
|
TTLL4
|
tubulin tyrosine ligase-like family, member 4
|
|
chr4_-_170924911
|
0.523
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr16_+_66914380
|
0.523
|
NM_020786
|
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2
|
|
chr2_+_70142152
|
0.523
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr4_+_78978723
|
0.520
|
NM_001166133
NM_025074
|
FRAS1
|
Fraser syndrome 1
|
|
chr6_+_106546736
|
0.516
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chrX_-_32430370
|
0.516
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr7_-_138665853
|
0.514
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr8_-_22550057
|
0.513
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr11_+_58939539
|
0.505
|
NM_015177
|
DTX4
|
deltex homolog 4 (Drosophila)
|
|
chr2_+_219187858
|
0.501
|
NM_022572
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chrX_-_32173585
|
0.500
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr8_-_22549901
|
0.500
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr11_+_59522531
|
0.496
|
NM_001178040
NM_004177
|
STX3
|
syntaxin 3
|
|
chr18_+_43914186
|
0.493
|
NM_152470
|
RNF165-IT1
RNF165
|
RNF165 intronic transcript 1 (non-protein coding)
ring finger protein 165
|
|
chr2_-_27435014
|
0.491
|
NM_021095
|
SLC5A6
|
solute carrier family 5 (sodium-dependent vitamin transporter), member 6
|
|
chr18_+_55714457
|
0.490
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_-_80656870
|
0.490
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr6_+_12290528
|
0.488
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chrX_-_31526414
|
0.488
|
NM_004014
|
DMD
|
dystrophin
|
|
chr7_+_90032647
|
0.487
|
NM_001185072
NM_001185073
NM_012129
|
CLDN12
|
claudin 12
|
|
chr16_+_57662301
|
0.484
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr11_+_44785975
|
0.480
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr3_-_190040192
|
0.478
|
NM_021101
|
CLDN1
|
claudin 1
|
|
chr16_+_18995255
|
0.475
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr1_-_204380903
|
0.473
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr2_+_75059781
|
0.471
|
NM_000189
|
HK2
|
hexokinase 2
|
|
chr5_+_148206155
|
0.468
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr9_-_126030854
|
0.467
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr3_-_185216545
|
0.467
|
NM_080652
|
TMEM41A
|
transmembrane protein 41A
|
|
chr8_-_17579729
|
0.466
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr16_+_57653909
|
0.463
|
NM_001145770
NM_005682
|
GPR56
|
G protein-coupled receptor 56
|
|
chr16_+_57673206
|
0.461
|
NM_001145774
|
GPR56
|
G protein-coupled receptor 56
|
|
chr8_-_17658385
|
0.453
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr8_-_17555158
|
0.451
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr9_-_140196702
|
0.446
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chrX_-_33229428
|
0.443
|
NM_004006
|
DMD
|
dystrophin
|
|
chr18_+_55711388
|
0.442
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr8_-_17580024
|
0.439
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chrX_-_117250624
|
0.437
|
NM_001168302
NM_001168303
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr3_-_52090090
|
0.436
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chrX_-_33146543
|
0.435
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr16_+_18995508
|
0.432
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr13_-_52585472
|
0.431
|
NM_000053
NM_001005918
NM_001243182
|
ATP7B
|
ATPase, Cu++ transporting, beta polypeptide
|
|
chr19_+_34287662
|
0.430
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chrX_-_117251302
|
0.426
|
NM_001168301
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr2_+_148602569
|
0.424
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr22_+_25960738
|
0.423
|
NM_005160
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr17_+_7342688
|
0.421
|
NM_004112
|
FGF11
|
fibroblast growth factor 11
|
|
chr11_-_118023534
|
0.417
|
NM_001142348
NM_174934
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr15_+_80987609
|
0.416
|
NM_021214
|
FAM108C1
|
family with sequence similarity 108, member C1
|
|
chr18_+_55816546
|
0.412
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_-_3456734
|
0.410
|
NM_015482
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chrX_+_16804531
|
0.409
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr14_-_53162418
|
0.406
|
NM_014584
|
ERO1L
|
ERO1-like (S. cerevisiae)
|
|
chr3_-_27498244
|
0.406
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr11_-_118016134
|
0.405
|
NM_001142349
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr18_+_55862577
|
0.403
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_+_71123105
|
0.399
|
NM_001105531
NM_020819
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chrX_-_117119282
|
0.398
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr2_+_182321618
|
0.394
|
NM_000885
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr2_-_230933618
|
0.391
|
NM_152527
|
SLC16A14
|
solute carrier family 16, member 14 (monocarboxylic acid transporter 14)
|
|
chr6_+_117198375
|
0.390
|
NM_173560
|
RFX6
|
regulatory factor X, 6
|
|
chr3_-_48229800
|
0.390
|
NM_001789
NM_201567
|
CDC25A
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr9_-_126030792
|
0.388
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr15_+_75494196
|
0.385
|
NM_015492
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr3_-_33700932
|
0.382
|
NM_001207044
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chrX_-_117107700
|
0.380
|
NM_033495
NM_001168300
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr7_-_36406446
|
0.378
|
NM_001199707
NM_001199706
NM_015314
NM_001199708
|
KIAA0895
|
KIAA0895
|
|
chr10_+_101419169
|
0.373
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr12_-_57472573
|
0.367
|
NM_001130963
NM_015257
|
TMEM194A
|
transmembrane protein 194A
|
|
chr7_-_36429695
|
0.365
|
NM_001100425
|
KIAA0895
|
KIAA0895
|
|
chr2_-_74669055
|
0.362
|
NM_001015055
|
RTKN
|
rhotekin
|
|
chr15_+_90544608
|
0.362
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chr11_-_16497917
|
0.361
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr20_+_49348050
|
0.360
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr17_+_3539761
|
0.353
|
NM_001031681
NM_004937
|
CTNS
|
cystinosin, lysosomal cystine transporter
|
|
chr2_+_191002485
|
0.349
|
NM_001042521
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr17_-_38721701
|
0.346
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chrX_+_105066531
|
0.344
|
NM_198465
|
NRK
|
Nik related kinase
|
|
chr3_-_33759684
|
0.343
|
NM_015097
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr15_+_73976621
|
0.343
|
NM_001024736
NM_025240
|
CD276
|
CD276 molecule
|
|
chr5_-_55008125
|
0.339
|
NM_173514
|
SLC38A9
|
solute carrier family 38, member 9
|
|
chr2_+_191045588
|
0.339
|
NM_001042519
NM_032321
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr16_-_17564737
|
0.338
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr7_-_71877359
|
0.337
|
NM_031468
|
CALN1
|
calneuron 1
|
|
chr11_-_16430425
|
0.337
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr8_-_144623611
|
0.334
|
NM_015117
|
ZC3H3
|
zinc finger CCCH-type containing 3
|
|
chr11_-_16424391
|
0.332
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr11_-_102668892
|
0.330
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr2_-_74781061
|
0.329
|
NM_032603
|
LOXL3
|
lysyl oxidase-like 3
|
|
chrX_+_152907892
|
0.329
|
NM_001395
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr2_+_191014971
|
0.328
|
NM_001042520
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr7_+_76090971
|
0.328
|
NM_001102594
NM_001102595
NM_020892
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr4_+_177241003
|
0.326
|
NM_021928
|
SPCS3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae)
|
|
chr16_+_22019438
|
0.323
|
NM_001164579
|
C16orf52
|
chromosome 16 open reading frame 52
|
|
chr3_-_53080038
|
0.322
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr16_+_81348564
|
0.320
|
NM_022041
|
GAN
|
gigaxonin
|
|
chr6_-_134373618
|
0.319
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr12_-_25055966
|
0.314
|
NM_001178093
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr22_+_21771661
|
0.312
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr22_+_50624353
|
0.312
|
NM_025204
|
TRABD
|
TraB domain containing
|
|
chr18_+_55888750
|
0.311
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chrX_+_134654546
|
0.308
|
NM_182540
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr16_+_70147528
|
0.308
|
NM_017990
|
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit
|
|
chr12_-_25102281
|
0.307
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr12_-_25055228
|
0.307
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr17_-_41836132
|
0.306
|
NM_025237
|
SOST
|
sclerostin
|
|
chr2_+_32390902
|
0.304
|
NM_001193513
NM_001193514
NM_001193515
NM_017964
|
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6
|
|
chrX_-_31284946
|
0.301
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chr9_+_101569933
|
0.299
|
NM_024642
|
GALNT12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12)
|
|
chr6_-_3445198
|
0.298
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr12_+_54366909
|
0.298
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr7_+_107221203
|
0.294
|
NM_001008405
|
BCAP29
|
B-cell receptor-associated protein 29
|
|
chrX_+_147582133
|
0.294
|
NM_001169122
NM_001169123
NM_001169124
NM_001169125
NM_002025
|
AFF2
|
AF4/FMR2 family, member 2
|
|
chr7_+_107220421
|
0.294
|
NM_018844
|
BCAP29
|
B-cell receptor-associated protein 29
|
|
chrX_+_118370205
|
0.292
|
NM_006667
|
PGRMC1
|
progesterone receptor membrane component 1
|
|
chr5_+_56110899
|
0.292
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr12_-_123451031
|
0.292
|
NM_001243013
NM_001243014
NM_019624
NM_019625
NM_203444
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr17_-_6459746
|
0.291
|
NM_001165966
NM_031220
|
PITPNM3
|
PITPNM family member 3
|
|
chr6_+_71136141
|
0.291
|
NM_001162529
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chr16_-_2908105
|
0.288
|
NM_022119
|
PRSS22
|
protease, serine, 22
|
|
chr8_+_86099909
|
0.288
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr14_-_95599794
|
0.287
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr4_+_74606222
|
0.286
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr2_+_153574385
|
0.285
|
NM_152522
|
ARL6IP6
|
ADP-ribosylation-like factor 6 interacting protein 6
|
|
chr1_+_32645322
|
0.284
|
NM_175852
|
TXLNA
|
taxilin alpha
|
|
chr20_+_9198036
|
0.284
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr2_+_86668270
|
0.283
|
NM_018433
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chrX_+_49832214
|
0.281
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr1_-_24126891
|
0.281
|
NM_000403
|
GALE
|
UDP-galactose-4-epimerase
|
|
chr22_+_29468887
|
0.279
|
NM_001039570
NM_032045
|
KREMEN1
|
kringle containing transmembrane protein 1
|
|
chr15_+_98503907
|
0.279
|
NM_183376
|
ARRDC4
|
arrestin domain containing 4
|
|
chr1_-_93646245
|
0.277
|
NM_001167830
NM_016040
|
TMED5
|
transmembrane emp24 protein transport domain containing 5
|
|
chr2_+_204801453
|
0.277
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chr17_+_29718641
|
0.276
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr17_+_1959512
|
0.276
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr5_-_175964223
|
0.273
|
NM_014901
|
RNF44
|
ring finger protein 44
|