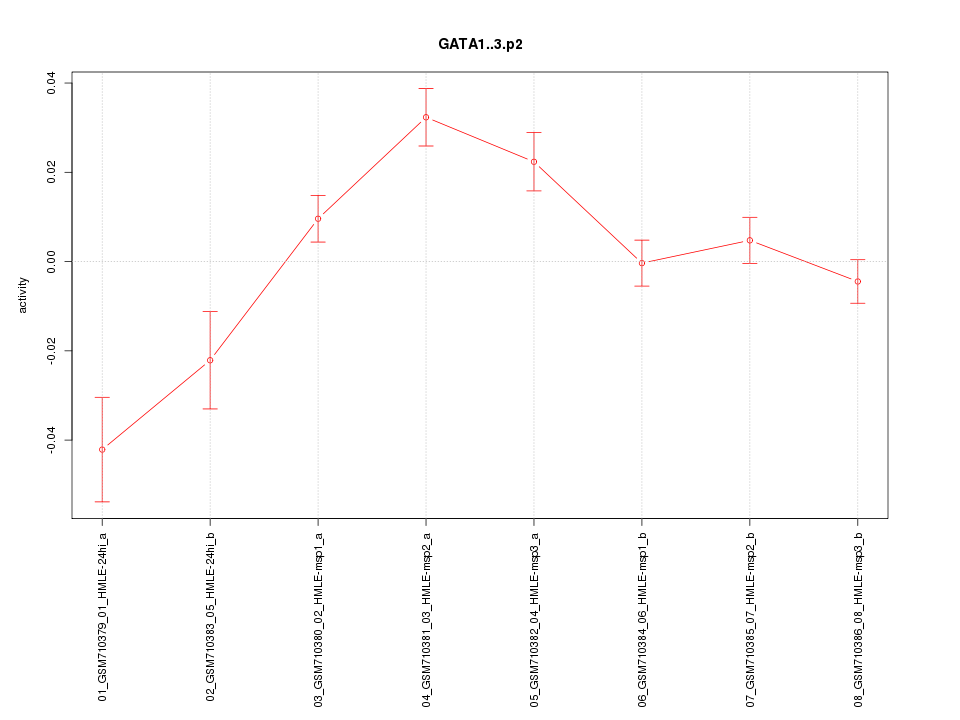
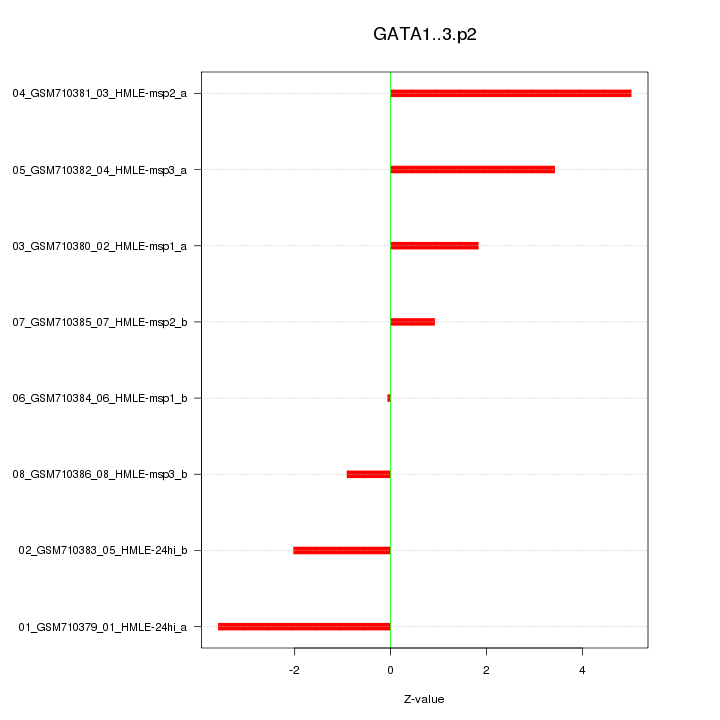
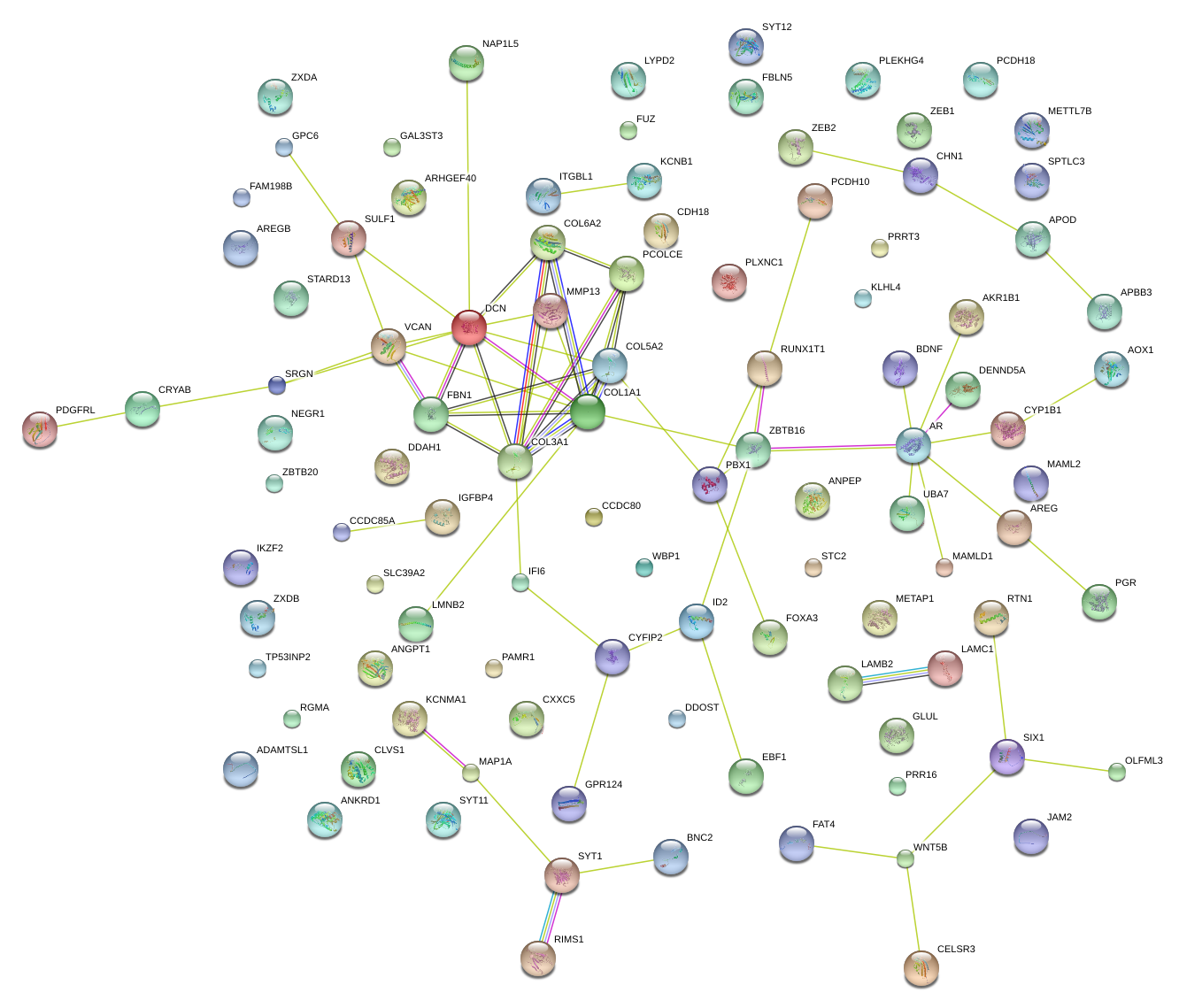
|
chr5_+_156820161
|
4.890
|
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr2_+_189839078
|
4.887
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189839132
|
4.426
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189839113
|
4.155
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr12_+_1738411
|
3.883
|
NM_032642
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr11_-_35547144
|
3.598
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr17_-_48278874
|
3.454
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr2_-_190044330
|
3.423
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr8_-_108510094
|
3.378
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chrX_+_57618268
|
3.289
|
NM_007157
|
ZXDB
|
zinc finger, X-linked, duplicated B
|
|
chr4_-_138453628
|
3.105
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr12_+_56075418
|
3.087
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr1_-_72748147
|
2.953
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr8_-_93107442
|
2.930
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_164528914
|
2.840
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr8_-_93107643
|
2.753
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr13_+_102104941
|
2.507
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr1_-_72748491
|
2.441
|
|
|
|
|
chr8_-_93029864
|
2.175
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr20_-_48099178
|
2.171
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr4_-_159092509
|
2.141
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr4_-_138453564
|
2.121
|
|
PCDH18
|
protocadherin 18
|
|
chr1_+_114522029
|
2.120
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chrX_-_57936853
|
2.103
|
NM_007156
|
ZXDA
|
zinc finger, X-linked, duplicated A
|
|
chr3_-_112358758
|
2.039
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr20_+_33299315
|
1.991
|
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr1_+_155829259
|
1.872
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr17_-_48264039
|
1.869
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr14_-_61115906
|
1.864
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr11_-_96076328
|
1.835
|
NM_032427
|
MAML2
|
mastermind-like 2 (Drosophila)
|
|
chr3_-_114343791
|
1.813
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr20_+_12989616
|
1.797
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chrX_+_57618440
|
1.789
|
|
ZXDB
|
zinc finger, X-linked, duplicated B
|
|
chr1_-_86043932
|
1.775
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_-_38303307
|
1.745
|
NM_000104
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chrX_+_86772714
|
1.729
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr14_-_92413738
|
1.697
|
|
FBLN5
|
fibulin 5
|
|
chr1_+_155853546
|
1.659
|
|
SYT11
|
synaptotagmin XI
|
|
chr3_-_114343052
|
1.651
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_+_38599651
|
1.625
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr8_+_37654757
|
1.623
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr3_-_195310985
|
1.621
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr15_-_93616358
|
1.613
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr5_+_119799898
|
1.601
|
|
PRR16
|
proline rich 16
|
|
chr2_-_38303265
|
1.589
|
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr3_-_48700317
|
1.589
|
NM_001407
|
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr15_-_93616308
|
1.583
|
|
RGMA
|
RGM domain family, member A
|
|
chr8_-_93107881
|
1.575
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr10_+_70847817
|
1.571
|
NM_002727
|
SRGN
|
serglycin
|
|
chr14_-_60097223
|
1.565
|
|
RTN1
|
reticulon 1
|
|
chr6_+_72596699
|
1.559
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr12_+_56075329
|
1.528
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr15_-_48937795
|
1.479
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr11_-_9225780
|
1.476
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr21_+_47545896
|
1.443
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr14_+_21538339
|
1.435
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chrX_+_66766350
|
1.433
|
|
AR
|
androgen receptor
|
|
chr5_-_139944045
|
1.433
|
NM_006051
NM_133172
NM_133173
NM_133174
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr10_+_70847857
|
1.416
|
|
SRGN
|
serglycin
|
|
chr5_-_19988293
|
1.411
|
NM_001167667
NM_004934
|
CDH18
|
cadherin 18, type 2
|
|
chr4_+_134073180
|
1.409
|
|
PCDH10
|
protocadherin 10
|
|
chr13_-_33780142
|
1.397
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr11_-_27742224
|
1.385
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_+_82767492
|
1.374
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr5_+_119800004
|
1.371
|
NM_016644
|
PRR16
|
proline rich 16
|
|
chr3_-_49170490
|
1.358
|
NM_002292
|
LAMB2
|
laminin, beta 2 (laminin S)
|
|
chr3_-_112358510
|
1.348
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr11_-_111782447
|
1.348
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr21_+_27012067
|
1.345
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr12_+_94542239
|
1.335
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr5_+_139060295
|
1.332
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr2_+_74685526
|
1.330
|
NM_012477
|
WBP1
|
WW domain binding protein 1
|
|
chr3_-_195310770
|
1.329
|
|
APOD
|
apolipoprotein D
|
|
chr2_-_214016332
|
1.329
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr5_-_158526698
|
1.327
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr4_-_89618928
|
1.326
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr3_-_49170427
|
1.307
|
|
LAMB2
|
laminin, beta 2 (laminin S)
|
|
chr13_+_93879011
|
1.305
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr9_+_18474078
|
1.303
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr12_-_91573264
|
1.300
|
NM_133503
|
DCN
|
decorin
|
|
chr10_-_92681025
|
1.297
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr12_-_91572328
|
1.291
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chrX_+_149531439
|
1.276
|
NM_001177465
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr10_-_79397204
|
1.269
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr2_-_175869910
|
1.269
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr3_-_49851356
|
1.260
|
NM_003335
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr3_-_49851217
|
1.255
|
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr15_-_93616891
|
1.247
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr16_+_67311412
|
1.246
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr11_-_65816650
|
1.240
|
NM_033036
|
GAL3ST3
|
galactose-3-O-sulfotransferase 3
|
|
chr9_-_16727887
|
1.231
|
|
BNC2
|
basonuclin 2
|
|
chr14_-_60097314
|
1.225
|
|
RTN1
|
reticulon 1
|
|
chr8_+_37654406
|
1.214
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr5_-_172755418
|
1.207
|
|
STC2
|
stanniocalcin 2
|
|
chr8_+_70405080
|
1.202
|
|
SULF1
|
sulfatase 1
|
|
chr1_-_182354649
|
1.201
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr8_+_62200524
|
1.195
|
NM_173519
|
CLVS1
|
clavesin 1
|
|
chr2_+_56411123
|
1.194
|
NM_001080433
|
CCDC85A
|
coiled-coil domain containing 85A
|
|
chr10_-_79397394
|
1.192
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr8_-_93075190
|
1.186
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_+_37654395
|
1.181
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr2_+_201473942
|
1.174
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr11_-_102826433
|
1.157
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr19_-_50316360
|
1.145
|
NM_001171937
NM_025129
|
FUZ
|
fuzzy homolog (Drosophila)
|
|
chr3_-_9994057
|
1.133
|
NM_207351
|
PRRT3
|
proline-rich transmembrane protein 3
|
|
chr15_-_90347534
|
1.133
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr10_-_79397290
|
1.119
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr14_+_21467413
|
1.112
|
NM_014579
|
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2
|
|
chr4_+_126237566
|
1.110
|
NM_024582
|
FAT4
|
FAT tumor suppressor homolog 4 (Drosophila)
|
|
chr8_+_17434700
|
1.109
|
|
PDGFRL
|
platelet-derived growth factor receptor-like
|
|
chr10_-_79397470
|
1.108
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr8_+_70378858
|
1.108
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr2_-_145277879
|
1.106
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr6_+_72926144
|
1.104
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr11_+_113930296
|
1.092
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr2_+_8822112
|
1.085
|
NM_002166
|
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein
|
|
chr1_-_27998700
|
1.074
|
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr7_+_100199881
|
1.060
|
NM_002593
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr11_-_2019007
|
1.056
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr19_+_46367517
|
1.055
|
NM_004497
|
FOXA3
|
forkhead box A3
|
|
chr12_+_79258794
|
1.055
|
|
SYT1
|
synaptotagmin I
|
|
chr15_-_93617091
|
1.054
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr13_-_33859835
|
1.049
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr15_+_43809805
|
1.043
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr8_+_37654833
|
1.040
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr10_-_104473957
|
1.037
|
NM_004311
|
ARL3
|
ADP-ribosylation factor-like 3
|
|
chr2_+_210444364
|
1.033
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr9_+_131191206
|
1.009
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr4_+_55095263
|
1.009
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr12_+_79258588
|
1.007
|
|
SYT1
|
synaptotagmin I
|
|
chr9_+_124086842
|
1.005
|
|
GSN
|
gelsolin
|
|
chr4_-_186877869
|
1.002
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_-_81205091
|
0.997
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr1_+_51435641
|
0.996
|
NM_078626
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr6_+_27777713
|
0.994
|
NM_003536
|
HIST1H3H
|
histone cluster 1, H3h
|
|
chr15_-_90347173
|
0.984
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr3_-_197676755
|
0.971
|
NM_001134435
|
IQCG
|
IQ motif containing G
|
|
chr3_-_51993955
|
0.960
|
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr17_-_48278987
|
0.933
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr19_-_1401474
|
0.928
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr17_-_5404304
|
0.928
|
NM_001162371
|
LOC728392
|
uncharacterized LOC728392
|
|
chr4_+_8951476
|
0.919
|
NM_001040071
|
LOC650293
|
seven transmembrane helix receptor
|
|
chr3_-_114478117
|
0.907
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr11_-_1769086
|
0.907
|
|
IFITM10
|
interferon induced transmembrane protein 10
|
|
chr2_+_210444154
|
0.904
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr3_+_45067750
|
0.901
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr10_-_92680784
|
0.900
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr19_-_50143350
|
0.894
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr9_+_35829208
|
0.893
|
NM_016446
NM_001042589
|
TMEM8B
|
transmembrane protein 8B
|
|
chr20_+_37353126
|
0.887
|
|
SLC32A1
|
solute carrier family 32 (GABA vesicular transporter), member 1
|
|
chr8_-_93111915
|
0.886
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_+_67584251
|
0.885
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_-_27998658
|
0.883
|
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr12_+_21679190
|
0.882
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr9_+_35829375
|
0.875
|
NM_001042590
|
TMEM8B
|
transmembrane protein 8B
|
|
chr18_-_34409084
|
0.868
|
NM_015476
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr2_-_145277568
|
0.864
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr11_+_62380212
|
0.859
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chrX_-_71792897
|
0.858
|
NM_001166418
NM_001166419
NM_001166420
NM_001166422
NM_001166448
NM_018486
|
HDAC8
|
histone deacetylase 8
|
|
chr10_+_69869249
|
0.857
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr1_+_51435911
|
0.855
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr2_+_27301693
|
0.853
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr6_+_72596648
|
0.850
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr5_-_168271852
|
0.849
|
|
|
|
|
chr10_-_79397546
|
0.848
|
NM_001014797
NM_001161352
NM_001161353
NM_002247
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr4_-_186733372
|
0.841
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr12_+_79258517
|
0.839
|
|
SYT1
|
synaptotagmin I
|
|
chr12_+_79258448
|
0.837
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr17_-_62050200
|
0.829
|
NM_000334
|
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit
|
|
chr2_-_127864505
|
0.825
|
NM_004305
NM_139343
NM_139344
NM_139345
NM_139346
NM_139347
NM_139348
NM_139349
NM_139350
NM_139351
|
BIN1
|
bridging integrator 1
|
|
chr4_+_129730772
|
0.823
|
NM_024900
NM_199320
|
PHF17
|
PHD finger protein 17
|
|
chr1_-_154943001
|
0.822
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr18_+_72998258
|
0.821
|
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr3_-_121379701
|
0.821
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr3_+_142838596
|
0.818
|
NM_004267
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2
|
|
chr18_-_34408845
|
0.816
|
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr20_+_37353104
|
0.815
|
NM_080552
|
SLC32A1
|
solute carrier family 32 (GABA vesicular transporter), member 1
|
|
chr3_-_9920633
|
0.811
|
NM_022094
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr17_-_48262900
|
0.809
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr19_+_49977817
|
0.806
|
NM_001204503
|
FLT3LG
|
fms-related tyrosine kinase 3 ligand
|
|
chr16_+_69680959
|
0.800
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr2_-_214015053
|
0.799
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr7_+_90338711
|
0.794
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr9_+_135037333
|
0.791
|
NM_032536
|
NTNG2
|
netrin G2
|
|
chr15_+_91769329
|
0.785
|
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr18_-_34408948
|
0.785
|
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr9_-_95244750
|
0.784
|
NM_001193335
NM_017680
|
ASPN
|
asporin
|
|
chr4_-_186732047
|
0.784
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_226016428
|
0.781
|
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr5_-_139943829
|
0.780
|
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr14_-_75593623
|
0.771
|
|
NEK9
|
NIMA (never in mitosis gene a)- related kinase 9
|
|
chr14_-_60097531
|
0.766
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr6_+_132891460
|
0.763
|
NM_175067
|
TAAR6
|
trace amine associated receptor 6
|
|
chrX_+_100333835
|
0.759
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr11_+_113930430
|
0.754
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr11_-_2019070
|
0.748
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr11_-_2016743
|
0.747
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr8_+_70405027
|
0.743
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chrX_+_102883647
|
0.734
|
NM_001006640
|
TCEAL1
|
transcription elongation factor A (SII)-like 1
|
|
chr13_-_38172862
|
0.734
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|