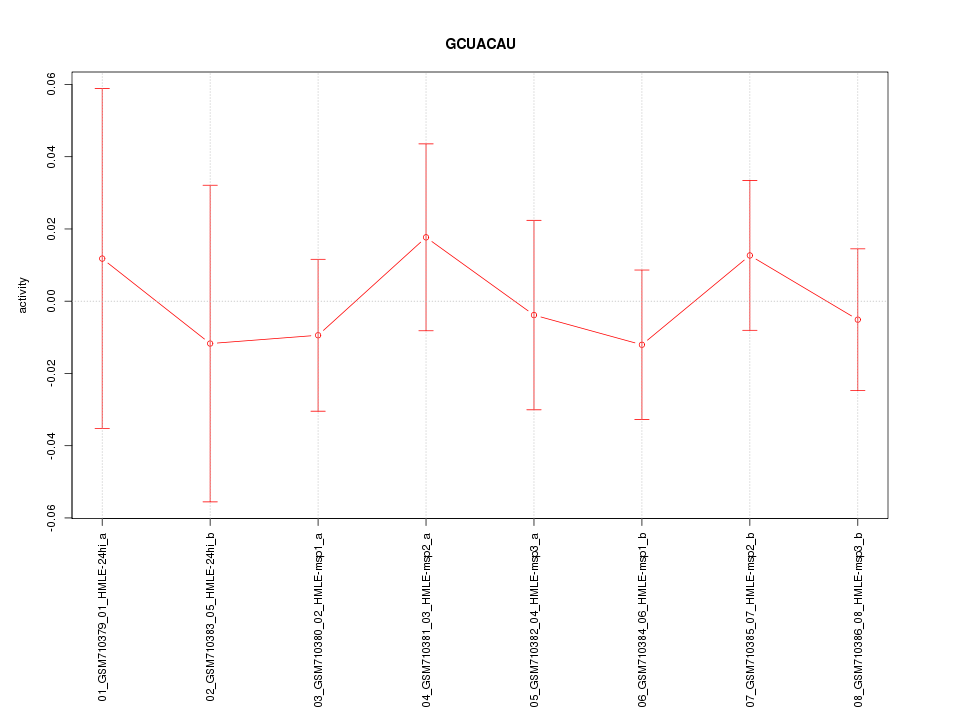
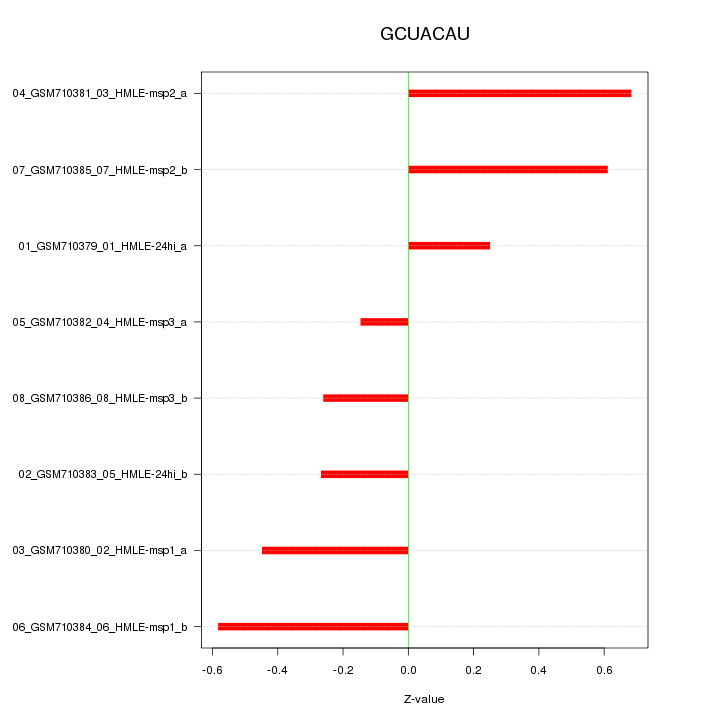
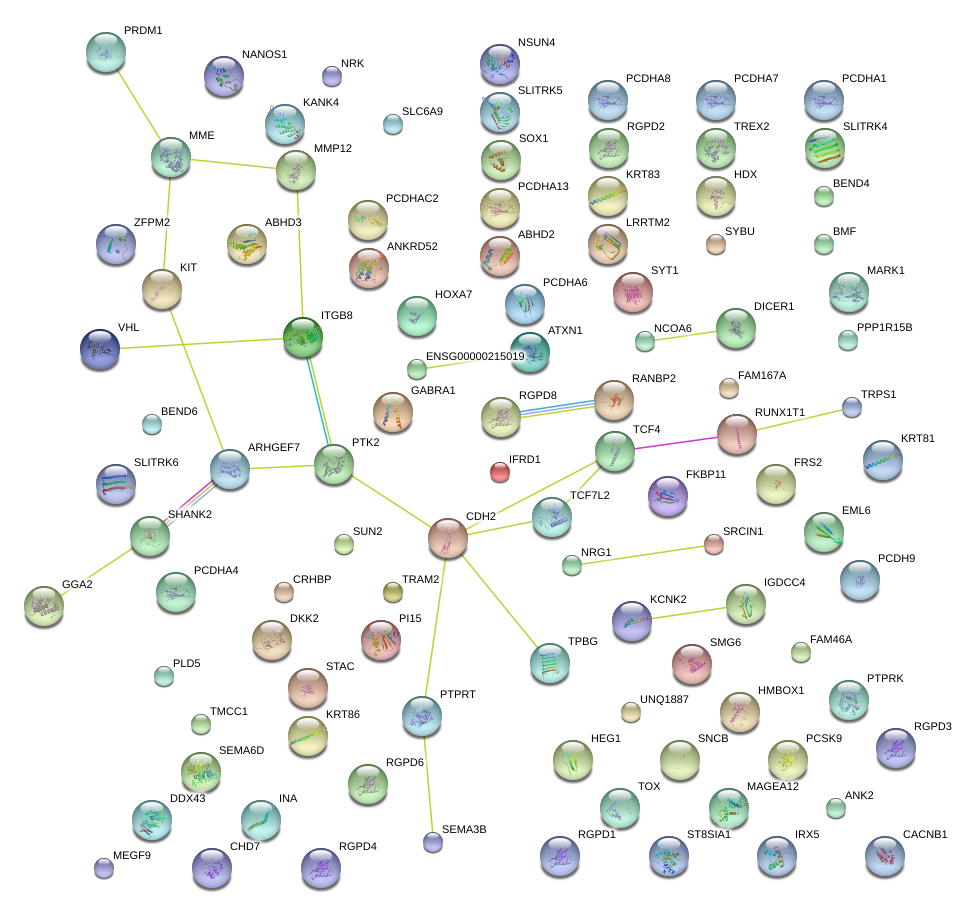
|
chr13_+_88324777
|
0.205
|
NM_015567
|
SLITRK5
|
SLIT and NTRK-like family, member 5
|
|
chr8_-_93115453
|
0.145
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_-_22487566
|
0.139
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr8_-_93107881
|
0.139
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93111915
|
0.138
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107442
|
0.134
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93029864
|
0.129
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93075190
|
0.123
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr4_+_113739203
|
0.117
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr4_+_113970784
|
0.107
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr8_+_32579350
|
0.104
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr1_-_62785067
|
0.096
|
NM_181712
|
KANK4
|
KN motif and ankyrin repeat domains 4
|
|
chr4_-_42154894
|
0.088
|
NM_001159547
NM_207406
|
BEND4
|
BEN domain containing 4
|
|
chr4_+_55523906
|
0.083
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr10_+_120789227
|
0.070
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr6_-_82462374
|
0.070
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr1_-_44497132
|
0.067
|
NM_001024845
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr6_-_16761600
|
0.062
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr1_-_44482996
|
0.061
|
NM_006934
NM_201649
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr8_-_116681103
|
0.059
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr9_-_123476578
|
0.055
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr6_-_52441815
|
0.052
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr5_+_161275541
|
0.046
|
NM_001127645
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr8_+_75736771
|
0.044
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr1_+_220701547
|
0.043
|
NM_018650
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr12_-_52685249
|
0.042
|
NM_002281
|
KRT81
|
keratin 81
|
|
chr5_+_161277391
|
0.041
|
NM_001127647
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_+_161274939
|
0.040
|
NM_001127644
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_+_161274684
|
0.040
|
NM_001127643
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr4_-_107957452
|
0.040
|
NM_014421
|
DKK2
|
dickkopf 2 homolog (Xenopus laevis)
|
|
chr1_+_215256559
|
0.040
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr12_-_121342050
|
0.039
|
NM_139015
|
SPPL3
|
signal peptide peptidase-like 3
|
|
chrX_-_83757398
|
0.038
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr8_+_28748030
|
0.038
|
NM_024567
|
HMBOX1
|
homeobox containing 1
|
|
chr13_+_111767586
|
0.037
|
NM_001113511
NM_001113512
NM_145735
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr8_+_32405727
|
0.037
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr1_+_215178884
|
0.036
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr5_+_161275897
|
0.036
|
NM_001127646
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_+_161274196
|
0.036
|
NM_000806
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr12_+_79257772
|
0.036
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr5_+_76248679
|
0.035
|
NM_001882
|
CRHBP
|
corticotropin releasing hormone binding protein
|
|
chr13_-_67804432
|
0.035
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr12_+_79439432
|
0.034
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr8_+_28747885
|
0.034
|
NM_001135726
|
HMBOX1
|
homeobox containing 1
|
|
chr18_-_25757351
|
0.034
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr7_-_27196163
|
0.034
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr8_-_110657025
|
0.034
|
NM_001099753
NM_001099754
NM_001099755
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr12_+_79258448
|
0.034
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr8_-_110704019
|
0.034
|
NM_001099743
NM_001099744
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr5_+_140220811
|
0.033
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr8_+_106331146
|
0.033
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr3_+_10183318
|
0.033
|
NM_000551
NM_198156
|
VHL
|
von Hippel-Lindau tumor suppressor
|
|
chr8_-_110661007
|
0.033
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr8_-_110693233
|
0.033
|
NM_001099745
NM_001099746
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chrX_-_142722869
|
0.032
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr3_-_124774735
|
0.032
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr18_-_53255734
|
0.031
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr7_+_20370724
|
0.031
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr8_-_110657745
|
0.031
|
NM_001099751
NM_001099752
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr8_-_60031545
|
0.031
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr16_-_23521803
|
0.030
|
NM_015044
|
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2
|
|
chrX_-_142722318
|
0.030
|
NM_001184750
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr5_+_140186658
|
0.030
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr8_-_110655418
|
0.030
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr17_-_36762105
|
0.030
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr5_-_138211054
|
0.030
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr5_+_140254930
|
0.030
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr15_+_47476225
|
0.029
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr5_+_140213968
|
0.029
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr12_-_56652118
|
0.029
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr5_+_140207562
|
0.029
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr5_+_140247767
|
0.029
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr1_+_46806389
|
0.027
|
NM_199044
|
NSUN4
|
NOP2/Sun domain family, member 4
|
|
chr6_+_83073954
|
0.027
|
NM_001166392
|
TPBG
|
trophoblast glycoprotein
|
|
chr2_+_54952148
|
0.027
|
NM_001039753
|
EML6
|
echinoderm microtubule associated protein like 6
|
|
chr22_-_39152008
|
0.027
|
NM_001199579
NM_015374
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr7_+_112090463
|
0.027
|
NM_001550
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr6_-_128841432
|
0.027
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr15_-_52970808
|
0.026
|
NM_019600
|
FAM214A
|
family with sequence similarity 214, member A
|
|
chr13_-_86373328
|
0.026
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr1_-_204380903
|
0.026
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr6_+_74104284
|
0.026
|
NM_018665
|
DDX43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43
|
|
chrX_-_142723921
|
0.025
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr18_-_19284737
|
0.025
|
NM_138340
|
ABHD3
|
abhydrolase domain containing 3
|
|
chr8_-_11324184
|
0.024
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr6_+_106534188
|
0.024
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr1_-_242687997
|
0.023
|
NM_152666
|
PLD5
|
phospholipase D family, member 5
|
|
chr6_+_56819706
|
0.023
|
NM_152731
|
BEND6
|
BEN domain containing 6
|
|
chr15_-_65715385
|
0.022
|
NM_020962
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4
|
|
chr3_+_36421944
|
0.022
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr15_+_89631380
|
0.022
|
NM_007011
NM_152924
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr1_+_55505148
|
0.022
|
NM_174936
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9
|
|
chr7_+_112092112
|
0.022
|
NM_001197079
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr3_-_129612371
|
0.022
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr1_-_242612757
|
0.021
|
NM_001195811
NM_001195812
|
PLD5
|
phospholipase D family, member 5
|
|
chr3_+_154798078
|
0.021
|
NM_007289
|
MME
|
membrane metallo-endopeptidase
|
|
chr3_+_154797644
|
0.021
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr22_-_39151457
|
0.021
|
NM_001199580
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr5_+_140165875
|
0.021
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr16_+_54965110
|
0.021
|
NM_001252197
NM_005853
|
IRX5
|
iroquois homeobox 5
|
|
chr14_-_95623665
|
0.021
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr20_-_41818464
|
0.021
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr3_+_50304989
|
0.021
|
NM_001005914
NM_004636
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr13_+_112721912
|
0.020
|
NM_005986
|
SOX1
|
SRY (sex determining region Y)-box 1
|
|
chr3_+_154797435
|
0.020
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr3_+_154797910
|
0.020
|
NM_007288
|
MME
|
membrane metallo-endopeptidase
|
|
chr5_-_176057324
|
0.020
|
NM_001001502
NM_003085
|
SNCB
|
synuclein, beta
|
|
chr2_+_109335906
|
0.020
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr5_+_140235594
|
0.020
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr5_+_140345746
|
0.020
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr10_+_105036783
|
0.019
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chrX_+_105066531
|
0.019
|
NM_198465
|
NRK
|
Nik related kinase
|
|
chr8_+_61591239
|
0.019
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr17_-_2206811
|
0.019
|
NM_001170957
|
SMG6
|
smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans)
|
|
chr11_-_70507911
|
0.019
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr12_+_69864065
|
0.019
|
NM_001042555
NM_006654
|
FRS2
|
fibroblast growth factor receptor substrate 2
|
|
chr15_-_40401041
|
0.019
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr6_+_106546736
|
0.019
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr8_-_142010987
|
0.019
|
NM_001199649
NM_005607
NM_153831
|
PTK2
|
PTK2 protein tyrosine kinase 2
|
|
chr16_-_18812691
|
0.018
|
NM_015161
|
ARL6IP1
|
ADP-ribosylation factor-like 6 interacting protein 1
|
|
chr11_-_16430425
|
0.018
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr2_+_201170795
|
0.018
|
NM_001100422
NM_001100424
NM_001100423
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr14_+_72398816
|
0.018
|
NM_001204423
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr11_-_70935807
|
0.018
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr5_+_161277702
|
0.018
|
NM_001127648
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr4_+_106816593
|
0.018
|
NM_001033047
NM_001184690
NM_001184691
NM_001184692
NM_001184693
|
NPNT
|
nephronectin
|
|
chr2_+_201170603
|
0.018
|
NM_015535
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr9_+_36036900
|
0.018
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr2_+_70142152
|
0.018
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr22_+_33196801
|
0.017
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr18_+_43914186
|
0.017
|
NM_152470
|
RNF165-IT1
RNF165
|
RNF165 intronic transcript 1 (non-protein coding)
ring finger protein 165
|
|
chr6_-_161694992
|
0.017
|
NM_020133
|
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta)
|
|
chr14_+_72399149
|
0.017
|
NM_001204424
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr3_+_169684549
|
0.017
|
NM_003262
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr7_+_130126035
|
0.017
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr4_+_52709165
|
0.017
|
NM_001040402
NM_015115
|
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr2_+_5832778
|
0.017
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr10_-_128077072
|
0.017
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr11_-_16497917
|
0.016
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr1_+_28585959
|
0.016
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr7_+_130131921
|
0.016
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr15_-_40398573
|
0.016
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr5_+_79703792
|
0.016
|
NM_001105251
NM_014733
|
ZFYVE16
|
zinc finger, FYVE domain containing 16
|
|
chr1_-_224033673
|
0.016
|
NM_001031685
NM_005426
|
TP53BP2
|
tumor protein p53 binding protein, 2
|
|
chr8_-_62627167
|
0.016
|
NM_001164754
NM_001164755
NM_001164756
NM_004318
NM_032466
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr10_-_73611050
|
0.016
|
NM_001042465
NM_001042466
NM_002778
|
PSAP
|
prosaposin
|
|
chr20_+_33814538
|
0.016
|
NM_006690
|
MMP24
|
matrix metallopeptidase 24 (membrane-inserted)
|
|
chr10_+_114709963
|
0.016
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr14_+_73603140
|
0.016
|
NM_000021
NM_007318
|
PSEN1
|
presenilin 1
|
|
chr12_+_113229548
|
0.015
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr5_-_150948504
|
0.015
|
NM_001447
|
FAT2
|
FAT tumor suppressor homolog 2 (Drosophila)
|
|
chr3_+_16926451
|
0.015
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr7_+_130131172
|
0.015
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr16_+_22019438
|
0.015
|
NM_001164579
|
C16orf52
|
chromosome 16 open reading frame 52
|
|
chr6_-_56112315
|
0.015
|
NM_030820
|
COL21A1
|
collagen, type XXI, alpha 1
|
|
chr2_-_38303307
|
0.015
|
NM_000104
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr3_-_129599220
|
0.015
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr6_-_88875766
|
0.014
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr4_-_100867868
|
0.014
|
NM_001031723
|
DNAJB14
|
DnaJ (Hsp40) homolog, subfamily B, member 14
|
|
chr16_-_69419833
|
0.014
|
NM_005652
|
TERF2
|
telomeric repeat binding factor 2
|
|
chr6_+_83072922
|
0.014
|
NM_006670
|
TPBG
|
trophoblast glycoprotein
|
|
chr2_-_100106478
|
0.014
|
NM_001037872
NM_016316
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr3_-_157823815
|
0.014
|
NM_001163678
NM_003030
NM_006884
|
SHOX2
|
short stature homeobox 2
|
|
chr4_+_56262051
|
0.014
|
NM_018475
|
TMEM165
|
transmembrane protein 165
|
|
chr12_+_41582167
|
0.013
|
NM_001164595
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr16_+_69599868
|
0.013
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr7_+_112063131
|
0.013
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr20_+_44330654
|
0.013
|
NM_172005
|
WFDC13
|
WAP four-disulfide core domain 13
|
|
chr1_-_160617042
|
0.013
|
NM_003037
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr5_+_156887026
|
0.013
|
NM_001099287
NM_001172292
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr6_+_7107799
|
0.013
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr17_-_3794036
|
0.013
|
NM_172206
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha
|
|
chr6_+_36853639
|
0.013
|
NM_152734
|
C6orf89
|
chromosome 6 open reading frame 89
|
|
chr16_-_11350038
|
0.013
|
NM_003745
|
SOCS1
|
suppressor of cytokine signaling 1
|
|
chr4_+_72204769
|
0.013
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr15_+_86220267
|
0.013
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr1_-_165738106
|
0.013
|
NM_019026
|
TMCO1
|
transmembrane and coiled-coil domains 1
|
|
chr15_-_65809603
|
0.013
|
NM_197960
|
DPP8
|
dipeptidyl-peptidase 8
|
|
chr11_-_16424391
|
0.013
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr6_+_7107986
|
0.013
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr1_-_33815498
|
0.013
|
NM_004427
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr2_-_27886443
|
0.012
|
NM_014860
|
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like
|
|
chr1_-_33841142
|
0.012
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr3_-_184870727
|
0.012
|
NM_001025266
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chr10_+_74033676
|
0.012
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|
|
chr6_+_90539569
|
0.012
|
NM_001137667
NM_001137668
NM_012115
|
CASP8AP2
|
caspase 8 associated protein 2
|
|
chr1_-_68697997
|
0.012
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chrX_-_148713400
|
0.012
|
NM_001174092
NM_032508
|
TMEM185A
|
transmembrane protein 185A
|
|
chr7_+_16793320
|
0.012
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr1_+_32645322
|
0.012
|
NM_175852
|
TXLNA
|
taxilin alpha
|
|
chr12_-_123594974
|
0.012
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr3_+_3841079
|
0.012
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr16_+_55542909
|
0.012
|
NM_017839
|
LPCAT2
|
lysophosphatidylcholine acyltransferase 2
|
|
chr9_+_103791030
|
0.012
|
NM_207299
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr8_-_27472298
|
0.012
|
NM_001831
|
CLU
|
clusterin
|
|
chr4_-_89744154
|
0.012
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr7_+_94285620
|
0.012
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr7_+_94285676
|
0.012
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr2_-_88926863
|
0.012
|
NM_004836
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3
|
|
chr20_-_43438881
|
0.012
|
NM_001205317
NM_182970
|
RIMS4
|
regulating synaptic membrane exocytosis 4
|