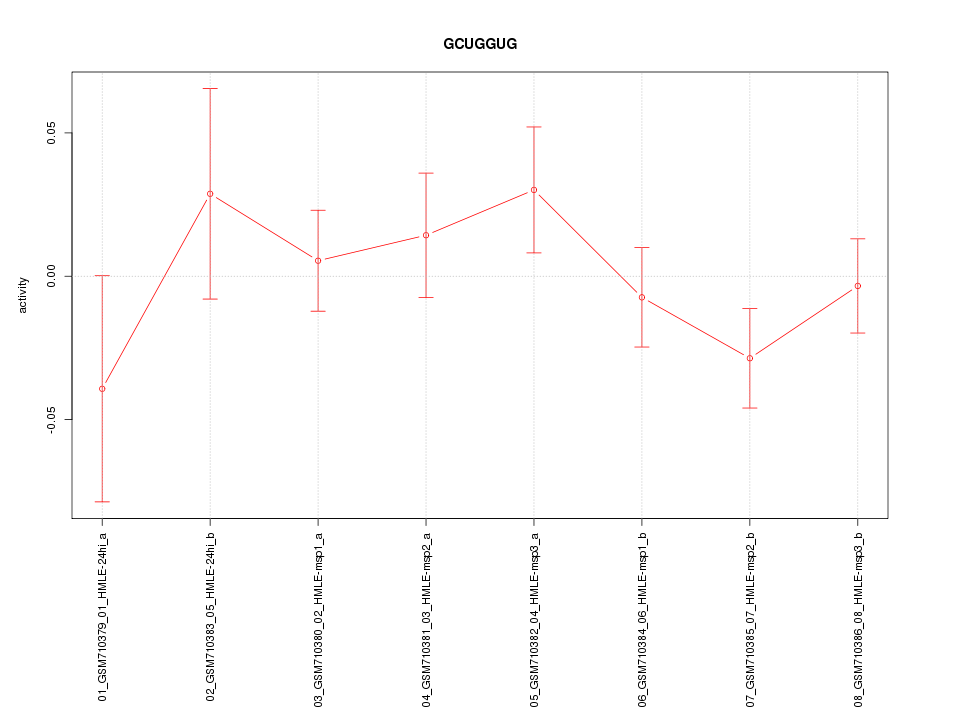
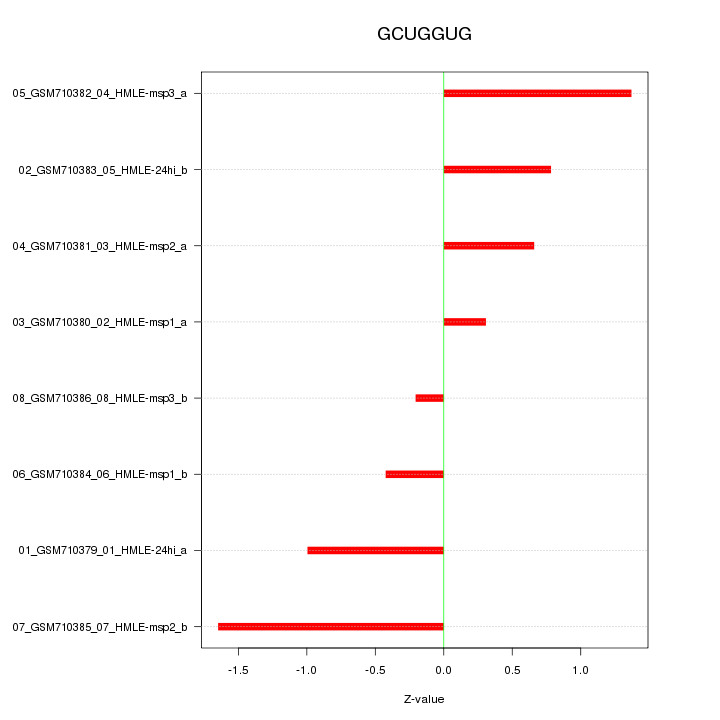
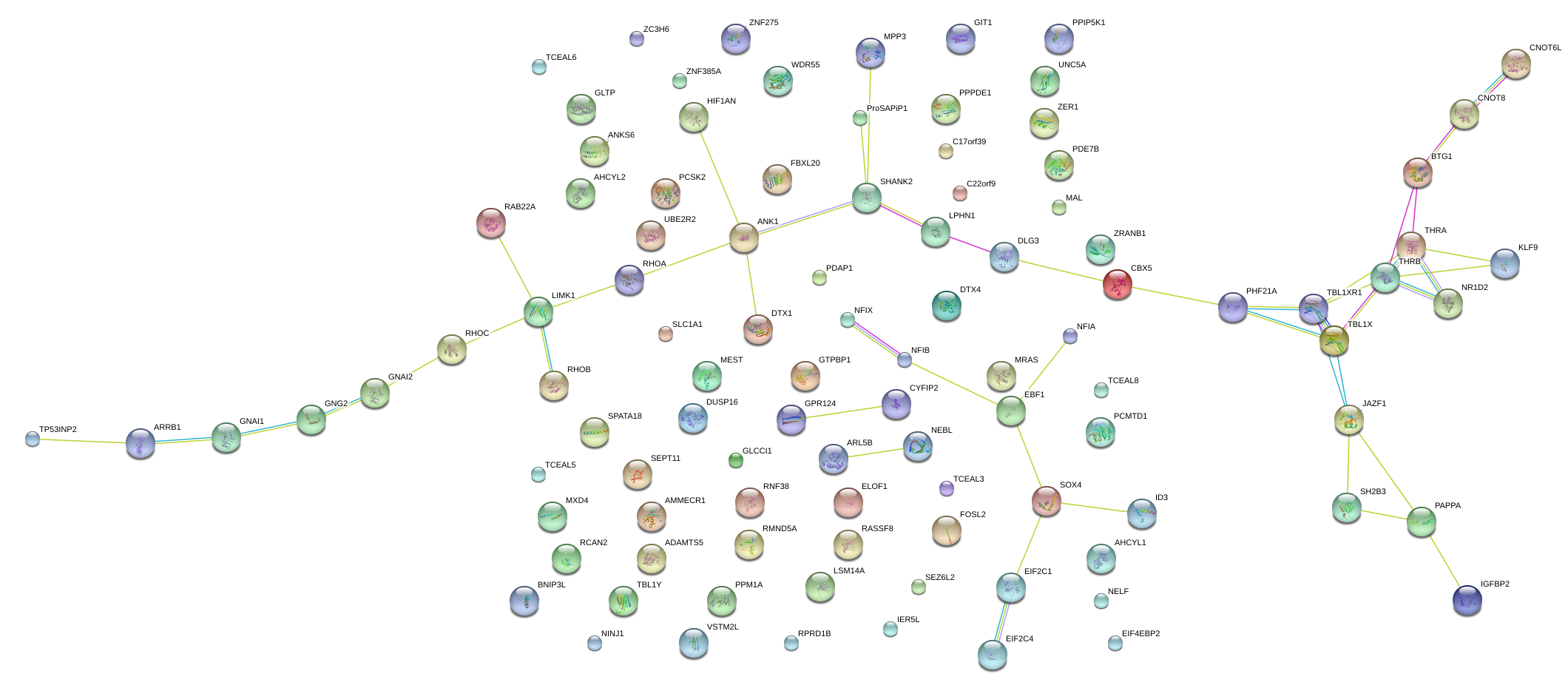
|
chr4_-_2263706
|
0.621
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr10_-_21463019
|
0.599
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr10_-_21186530
|
0.486
|
NM_006393
|
NEBL
|
nebulette
|
|
chr9_-_95896498
|
0.481
|
NM_004148
|
NINJ1
|
ninjurin 1
|
|
chr6_+_136172802
|
0.445
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr10_+_72163860
|
0.409
|
NM_004096
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr14_+_52327021
|
0.392
|
NM_001243773
NM_001243774
NM_053064
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr5_-_158526698
|
0.382
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr4_+_52917440
|
0.378
|
NM_145263
|
SPATA18
|
spermatogenesis associated 18 homolog (rat)
|
|
chr5_+_154238196
|
0.376
|
NM_004779
|
CNOT8
|
CCR4-NOT transcription complex, subunit 8
|
|
chr9_-_131534186
|
0.361
|
NM_006336
|
ZER1
|
zer-1 homolog (C. elegans)
|
|
chr11_+_58939539
|
0.360
|
NM_015177
|
DTX4
|
deltex homolog 4 (Drosophila)
|
|
chr20_+_33292117
|
0.358
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr12_-_92539614
|
0.341
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr4_-_78740508
|
0.332
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr9_-_131940538
|
0.331
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr20_+_36531498
|
0.327
|
NM_080607
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr2_+_113033146
|
0.323
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chrX_-_101397370
|
0.323
|
NM_001006938
|
TCEAL6
|
transcription elongation factor A (SII)-like 6
|
|
chr17_+_17942584
|
0.319
|
NM_024052
|
C17orf39
|
chromosome 17 open reading frame 39
|
|
chr6_-_46293628
|
0.319
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr14_+_60712469
|
0.317
|
NM_177952
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr8_-_41754244
|
0.310
|
NM_001142446
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr8_-_41655139
|
0.307
|
NM_000037
NM_020475
NM_020476
NM_020477
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chrX_-_102531673
|
0.306
|
NM_001012979
|
TCEAL5
|
transcription elongation factor A (SII)-like 5
|
|
chr9_+_118916069
|
0.303
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr8_+_26240522
|
0.300
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr8_-_41522724
|
0.295
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chrX_+_102862833
|
0.284
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chrX_+_152599544
|
0.271
|
NM_001080485
|
ZNF275
|
zinc finger protein 275
|
|
chr11_-_70935807
|
0.267
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr2_+_217498082
|
0.261
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr1_+_244816349
|
0.261
|
NM_016076
|
PPPDE1
|
PPPDE peptidase domain containing 1
|
|
chr7_+_129015483
|
0.260
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr5_+_156693051
|
0.260
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr2_+_86947387
|
0.260
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr11_-_70507911
|
0.257
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr7_-_28220342
|
0.255
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr7_+_8008416
|
0.247
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr7_+_73507485
|
0.231
|
NM_001204426
|
LIMK1
|
LIM domain kinase 1
|
|
chr9_-_140353785
|
0.231
|
NM_001130969
NM_001130970
NM_001130971
NM_001178064
NM_015537
|
NELF
|
nasal embryonic LHRH factor
|
|
chr11_-_46142955
|
0.228
|
NM_001101802
NM_016621
|
PHF21A
|
PHD finger protein 21A
|
|
chr3_+_23987514
|
0.227
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr12_+_111843743
|
0.227
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr5_+_156696351
|
0.227
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr19_+_13105970
|
0.227
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr7_+_128864740
|
0.225
|
NM_001130720
NM_015328
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr11_-_75062650
|
0.217
|
NM_004041
NM_020251
|
ARRB1
|
arrestin, beta 1
|
|
chr21_-_28339405
|
0.216
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr12_-_54778456
|
0.215
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr1_+_36273786
|
0.211
|
NM_017629
|
EIF2C4
|
eukaryotic translation initiation factor 2C, 4
|
|
chr5_+_176237435
|
0.208
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr1_-_23886284
|
0.208
|
NM_002167
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein
|
|
chr19_-_14316980
|
0.208
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr7_+_130126035
|
0.201
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr20_-_3149070
|
0.201
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr7_+_130131172
|
0.201
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr12_-_54785018
|
0.201
|
NM_001130967
NM_001130968
|
ZNF385A
|
zinc finger protein 385A
|
|
chr22_-_45636584
|
0.201
|
NM_001009880
|
KIAA0930
|
KIAA0930
|
|
chr3_+_23986735
|
0.200
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr22_-_45608346
|
0.199
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr8_+_37654395
|
0.190
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr5_+_140044383
|
0.187
|
NM_017706
|
WDR55
|
WD repeat domain 55
|
|
chr9_+_33817168
|
0.186
|
NM_017811
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr1_+_36348795
|
0.185
|
NM_012199
|
EIF2C1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr1_-_113249677
|
0.184
|
NM_001042679
|
RHOC
|
ras homolog gene family, member C
|
|
chr4_+_77870864
|
0.182
|
NM_018243
|
SEPT11
|
septin 11
|
|
chr7_+_130131921
|
0.181
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr8_-_52811707
|
0.180
|
NM_052937
|
PCMTD1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1
|
|
chr7_+_129007942
|
0.179
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr17_-_37557851
|
0.176
|
NM_001184906
NM_032875
|
FBXL20
|
F-box and leucine-rich repeat protein 20
|
|
chrX_-_102510048
|
0.175
|
NM_001006684
NM_153333
|
TCEAL8
|
transcription elongation factor A (SII)-like 8
|
|
chr20_+_17206751
|
0.175
|
NM_001201528
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr9_-_73029539
|
0.174
|
NM_001206
|
KLF9
|
Kruppel-like factor 9
|
|
chr3_-_24536176
|
0.173
|
NM_000461
NM_001128176
NM_001128177
NM_001252634
|
THRB
|
thyroid hormone receptor, beta
|
|
chr3_+_138066489
|
0.172
|
NM_001252091
NM_012219
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr19_+_34663311
|
0.171
|
NM_001114093
NM_015578
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr19_-_11670003
|
0.169
|
NM_032377
|
ELOF1
|
elongation factor 1 homolog (S. cerevisiae)
|
|
chr15_+_49170264
|
0.169
|
NM_014335
|
EID1
|
EP300 interacting inhibitor of differentiation 1
|
|
chr22_+_39101806
|
0.169
|
NM_004286
|
GTPBP1
|
GTP binding protein 1
|
|
chr1_+_61547625
|
0.168
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr1_-_113250022
|
0.167
|
NM_001042678
NM_175744
|
RHOC
|
ras homolog gene family, member C
|
|
chr2_+_28615771
|
0.165
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr7_+_73498106
|
0.165
|
NM_002314
|
LIMK1
|
LIM domain kinase 1
|
|
chr17_-_27916602
|
0.164
|
NM_001085454
NM_014030
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1
|
|
chr10_+_126630691
|
0.161
|
NM_017580
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1
|
|
chr20_+_54933982
|
0.160
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr7_-_99006275
|
0.155
|
NM_014891
|
PDAP1
|
PDGFA associated protein 1
|
|
chr10_+_18948276
|
0.153
|
NM_178815
|
ARL5B
|
ADP-ribosylation factor-like 5B
|
|
chr3_+_50284325
|
0.150
|
NM_001166425
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr6_+_21593963
|
0.148
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr10_+_102295572
|
0.145
|
NM_017902
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor
|
|
chr15_-_43877043
|
0.145
|
NM_014659
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1
|
|
chr20_+_17207596
|
0.145
|
NM_001201529
NM_002594
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr16_-_29910340
|
0.145
|
NM_001114099
NM_001114100
NM_001243332
NM_001243333
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chrX_+_9431314
|
0.144
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr1_+_61542928
|
0.140
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr12_-_110318178
|
0.140
|
NM_016433
|
GLTP
|
glycolipid transfer protein
|
|
chr12_-_12715244
|
0.139
|
NM_030640
|
DUSP16
|
dual specificity phosphatase 16
|
|
chr12_+_26126687
|
0.138
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chrX_+_69664704
|
0.135
|
NM_021120
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr12_-_54673901
|
0.135
|
NM_012117
|
CBX5
|
chromobox homolog 5
|
|
chr9_+_4490193
|
0.134
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr20_+_56884749
|
0.134
|
NM_020673
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr9_-_101558494
|
0.132
|
NM_173551
|
ANKS6
|
ankyrin repeat and sterile alpha motif domain containing 6
|
|
chr3_+_138067444
|
0.131
|
NM_001085049
NM_001252093
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr9_-_36400212
|
0.131
|
NM_022781
NM_194329
|
RNF38
|
ring finger protein 38
|
|
chr20_+_36661849
|
0.130
|
NM_021215
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chrX_-_109683457
|
0.129
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr12_+_113495661
|
0.128
|
NM_004416
|
DTX1
|
deltex homolog 1 (Drosophila)
|
|
chr5_-_114880536
|
0.127
|
NM_020177
|
FEM1C
|
fem-1 homolog c (C. elegans)
|
|
chr5_-_10761344
|
0.127
|
NM_004394
|
DAP
|
death-associated protein
|
|
chr2_-_213403280
|
0.127
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr16_+_1031807
|
0.127
|
NM_014587
|
SOX8
|
SRY (sex determining region Y)-box 8
|
|
chr10_-_60027630
|
0.126
|
NM_152230
|
IPMK
|
inositol polyphosphate multikinase
|
|
chr1_+_29213569
|
0.126
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr17_+_74864427
|
0.125
|
NM_001199172
NM_144677
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr1_-_27930101
|
0.125
|
NM_001029882
|
AHDC1
|
AT hook, DNA binding motif, containing 1
|
|
chr22_+_37415701
|
0.124
|
NM_001013436
NM_001130517
NM_021126
|
MPST
|
mercaptopyruvate sulfurtransferase
|
|
chr17_-_58603492
|
0.124
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr9_-_130679210
|
0.123
|
NM_175039
NM_175040
|
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4
|
|
chr12_+_19592311
|
0.123
|
NM_001114176
NM_153207
|
AEBP2
|
AE binding protein 2
|
|
chr20_+_306205
|
0.123
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr11_-_77348758
|
0.123
|
NM_001293
|
CLNS1A
|
chloride channel, nucleotide-sensitive, 1A
|
|
chr15_-_75199461
|
0.122
|
NM_020447
|
C15orf17
|
chromosome 15 open reading frame 17
|
|
chr1_+_44173217
|
0.122
|
NM_006279
NM_174963
NM_174964
NM_174965
NM_174966
NM_174967
NM_174968
NM_174969
NM_174970
NM_174971
|
ST3GAL3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3
|
|
chr4_-_186697065
|
0.121
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chrX_+_69672110
|
0.121
|
NM_020730
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr8_+_37887990
|
0.121
|
NM_004095
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1
|
|
chr6_-_111136407
|
0.120
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chrX_-_118827322
|
0.120
|
NM_015129
NM_145799
NM_145800
NM_145802
|
SEPT6
|
septin 6
|
|
chr12_+_26205490
|
0.120
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr11_-_130184459
|
0.120
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr7_+_140774031
|
0.118
|
NM_001195278
|
LOC100507421
|
transmembrane protein 178-like
|
|
chr2_+_219263060
|
0.117
|
NM_001206878
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr3_+_50273645
|
0.117
|
NM_002070
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr6_+_39760772
|
0.115
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr9_-_36401149
|
0.114
|
NM_194330
|
RNF38
|
ring finger protein 38
|
|
chr1_+_61547388
|
0.112
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr4_-_186732208
|
0.111
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr7_-_30029284
|
0.111
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr4_-_186877869
|
0.111
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr12_+_69864065
|
0.111
|
NM_001042555
NM_006654
|
FRS2
|
fibroblast growth factor receptor substrate 2
|
|
chr10_+_74451838
|
0.110
|
NM_138357
|
MCU
|
mitochondrial calcium uniporter
|
|
chr17_+_7608413
|
0.109
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr11_-_57417247
|
0.108
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr10_+_112836784
|
0.107
|
NM_000681
|
ADRA2A
|
adrenergic, alpha-2A-, receptor
|
|
chr7_+_90338711
|
0.107
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr15_+_52043738
|
0.107
|
NM_001142885
NM_014548
|
TMOD2
|
tropomodulin 2 (neuronal)
|
|
chr4_-_186732047
|
0.106
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_+_39760148
|
0.106
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr12_-_54653317
|
0.106
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chrX_+_72223351
|
0.105
|
NM_001042506
|
PABPC1L2B
|
poly(A) binding protein, cytoplasmic 1-like 2B
|
|
chr1_+_40840304
|
0.104
|
NM_001198978
|
SMAP2
|
small ArfGAP2
|
|
chr9_-_36400817
|
0.104
|
NM_194328
NM_194332
|
RNF38
|
ring finger protein 38
|
|
chr4_-_140060601
|
0.104
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr1_+_40859016
|
0.103
|
NM_001198979
|
SMAP2
|
small ArfGAP2
|
|
chr4_-_186733372
|
0.103
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_+_107811272
|
0.102
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr18_+_46065367
|
0.101
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr3_+_140950671
|
0.101
|
NM_001037172
NM_152282
|
ACPL2
|
acid phosphatase-like 2
|
|
chr1_+_40862471
|
0.101
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr12_-_54653369
|
0.100
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr2_+_219264477
|
0.099
|
NM_021198
NM_182642
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chrX_-_111326003
|
0.098
|
NM_012471
|
TRPC5
|
transient receptor potential cation channel, subfamily C, member 5
|
|
chr15_+_66161769
|
0.098
|
NM_001206836
NM_004663
|
RAB11A
|
RAB11A, member RAS oncogene family
|
|
chr11_+_66234293
|
0.098
|
NM_001098510
NM_001243136
NM_145065
|
PELI3
|
pellino homolog 3 (Drosophila)
|
|
chr7_-_30029904
|
0.097
|
NM_001145514
|
SCRN1
|
secernin 1
|
|
chr19_+_12863406
|
0.097
|
NM_017682
|
BEST2
|
bestrophin 2
|
|
chrX_+_69674914
|
0.096
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr5_-_150537335
|
0.095
|
NM_001155
|
ANXA6
|
annexin A6
|
|
chr9_+_37800779
|
0.094
|
NM_024345
|
DCAF10
|
DDB1 and CUL4 associated factor 10
|
|
chr13_-_67804432
|
0.094
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr2_+_74756531
|
0.094
|
NM_013247
NM_145074
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr5_-_150521200
|
0.094
|
NM_001193544
|
ANXA6
|
annexin A6
|
|
chr13_-_74707913
|
0.091
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr1_-_153895450
|
0.091
|
NM_020699
|
GATAD2B
|
GATA zinc finger domain containing 2B
|
|
chr22_+_46546498
|
0.090
|
NM_001001928
NM_005036
|
PPARA
|
peroxisome proliferator-activated receptor alpha
|
|
chr22_+_39052628
|
0.089
|
NM_001002880
NM_015373
|
CBY1
|
chibby homolog 1 (Drosophila)
|
|
chr1_+_204042191
|
0.089
|
NM_005686
|
SOX13
|
SRY (sex determining region Y)-box 13
|
|
chr20_+_32077904
|
0.088
|
NM_001032999
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr19_-_8070468
|
0.088
|
NM_001419
|
ELAVL1
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R)
|
|
chr2_-_145277879
|
0.087
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr10_+_181410
|
0.086
|
NM_001202464
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr1_+_16693582
|
0.085
|
NM_001114600
NM_015609
|
C1orf144
|
chromosome 1 open reading frame 144
|
|
chrX_-_109561314
|
0.083
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr17_-_1395950
|
0.082
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr3_+_38495778
|
0.081
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr8_-_135708789
|
0.080
|
NM_001029939
NM_001167583
NM_001174158
|
ZFAT
|
zinc finger and AT hook domain containing
|
|
chr2_+_135809834
|
0.080
|
NM_001172435
NM_012233
|
RAB3GAP1
|
RAB3 GTPase activating protein subunit 1 (catalytic)
|
|
chr2_+_85581840
|
0.080
|
NM_001135021
NM_001135022
NM_001135023
NM_032213
|
ELMOD3
|
ELMO/CED-12 domain containing 3
|
|
chr2_+_109237679
|
0.080
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr20_-_48732479
|
0.079
|
NM_022442
NM_021988
NM_199144
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr6_-_39895317
|
0.078
|
NM_005943
|
MOCS1
|
molybdenum cofactor synthesis 1
|
|
chr22_-_39240016
|
0.078
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr1_-_31381436
|
0.077
|
NM_014654
|
SDC3
|
syndecan 3
|
|
chr9_+_115513050
|
0.077
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr17_+_36861857
|
0.077
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr17_-_4890664
|
0.076
|
NM_001171167
NM_001171168
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr15_-_43882387
|
0.076
|
NM_001130858
NM_001130859
NM_001190214
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1
|