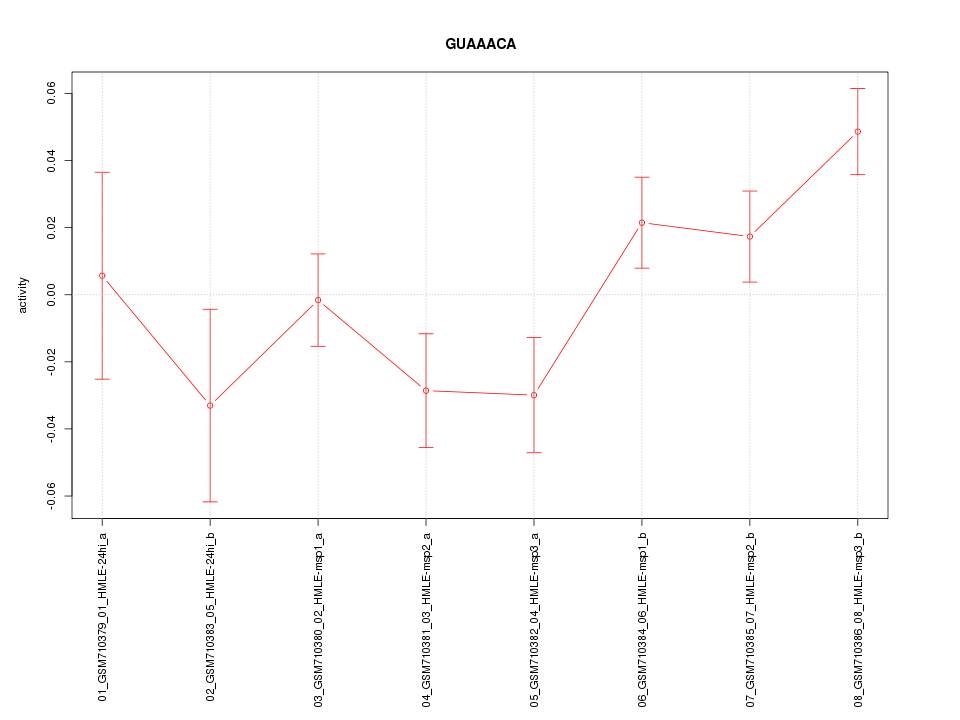
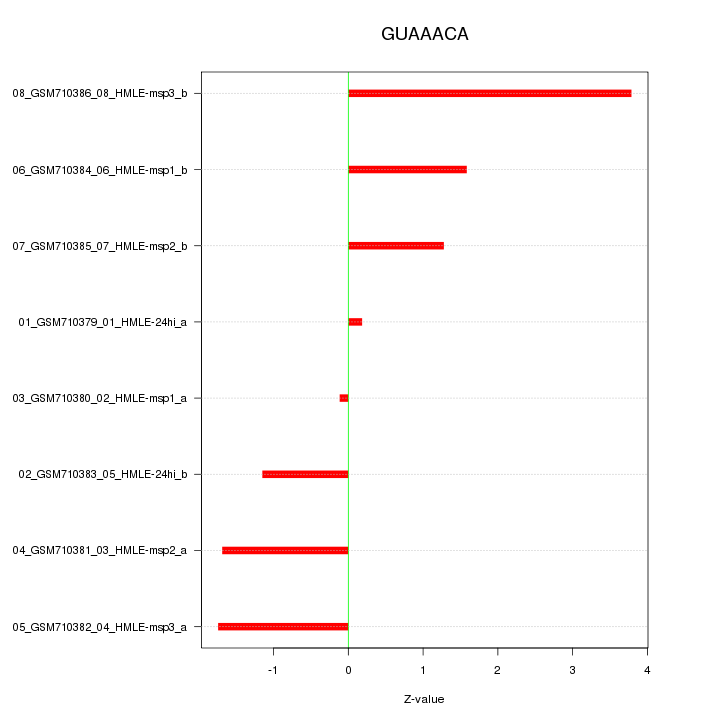
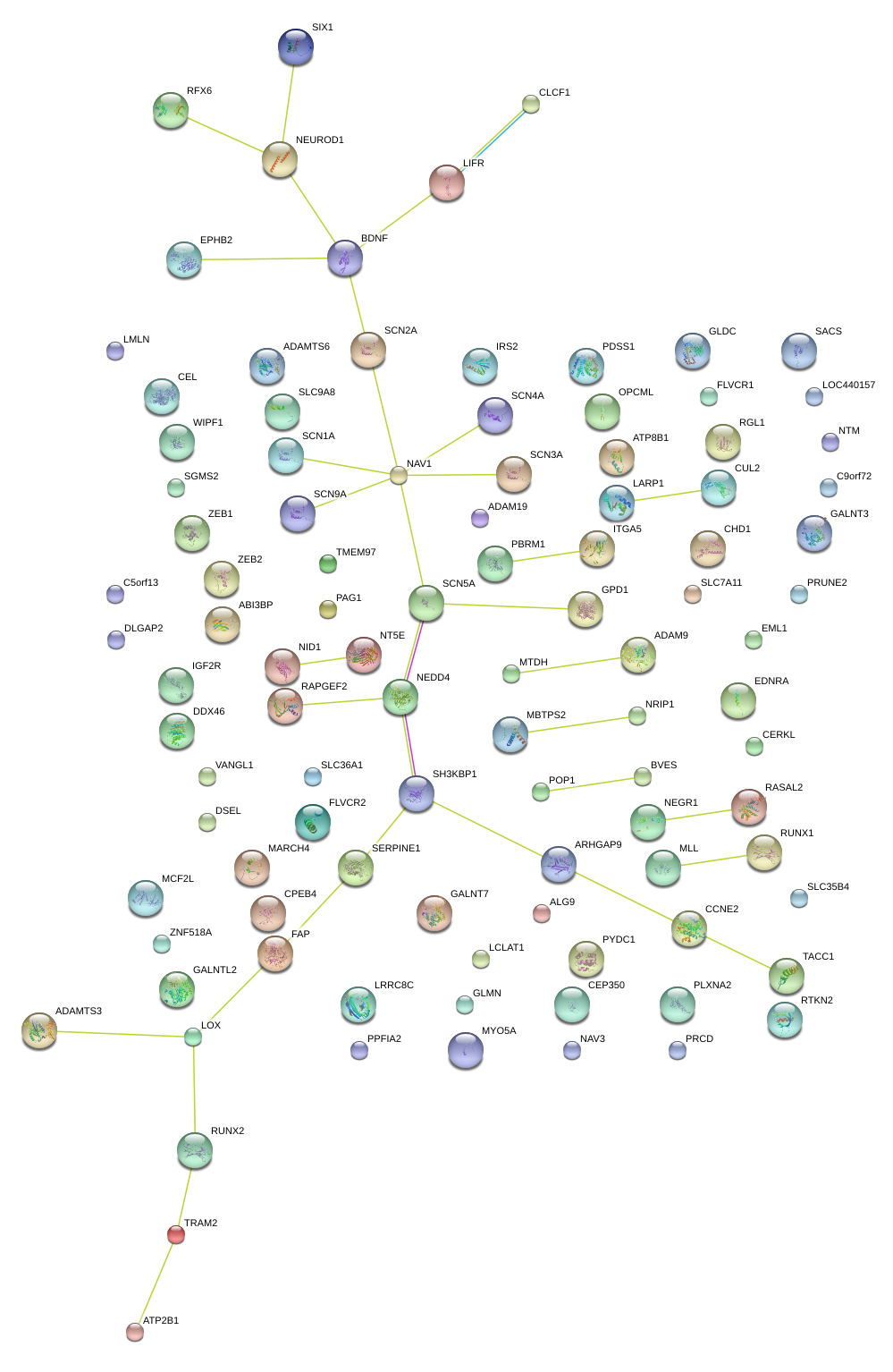
|
chr12_+_78225068
|
0.973
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr1_-_236228476
|
0.712
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr4_+_148402068
|
0.671
|
NM_001166055
NM_001957
|
EDNRA
|
endothelin receptor type A
|
|
chr5_-_157002739
|
0.659
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr14_-_61115906
|
0.640
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr1_-_208417635
|
0.629
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr1_-_72748147
|
0.628
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr8_-_95907423
|
0.627
|
NM_057749
|
CCNE2
|
cyclin E2
|
|
chr11_+_131240370
|
0.606
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr1_+_23037329
|
0.599
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr11_-_27743604
|
0.577
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27681195
|
0.574
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_+_179923870
|
0.567
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr11_-_27741192
|
0.563
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27721975
|
0.559
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27723061
|
0.555
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27721179
|
0.553
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_-_111093251
|
0.548
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr13_-_24007822
|
0.548
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr2_-_163099877
|
0.548
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr18_-_55470279
|
0.537
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr14_+_76071804
|
0.529
|
NM_001195283
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr5_-_121414011
|
0.529
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr5_-_111092918
|
0.524
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr11_-_27722446
|
0.523
|
NM_001143812
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr14_+_76044939
|
0.518
|
NM_017791
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr2_-_167232464
|
0.516
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr5_-_121412805
|
0.516
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr11_-_27742224
|
0.515
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_-_111093030
|
0.511
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_+_86159301
|
0.510
|
NM_001204813
NM_002526
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr11_-_27722599
|
0.508
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_+_201617449
|
0.492
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr1_+_116184573
|
0.478
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr5_-_111091947
|
0.476
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr14_+_100259445
|
0.474
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr7_+_100770378
|
0.472
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr3_+_197687070
|
0.468
|
NM_001136049
NM_033029
|
LMLN
|
leishmanolysin-like (metallopeptidase M8 family)
|
|
chr15_-_52821004
|
0.461
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr5_-_111093792
|
0.447
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111312522
|
0.445
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_213031596
|
0.443
|
NM_014053
|
FLVCR1
|
feline leukemia virus subgroup C cellular receptor 1
|
|
chr1_+_183605207
|
0.440
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr11_-_111741980
|
0.439
|
NM_001077691
NM_001077692
|
ALG9
|
asparagine-linked glycosylation 9, alpha-1,2-mannosyltransferase homolog (S. cerevisiae)
|
|
chr1_+_90098643
|
0.434
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr5_-_64777703
|
0.434
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr11_-_111742205
|
0.426
|
NM_001077690
NM_024740
|
ALG9
|
asparagine-linked glycosylation 9, alpha-1,2-mannosyltransferase homolog (S. cerevisiae)
|
|
chr1_+_116185249
|
0.423
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr13_-_110438896
|
0.420
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr11_-_67141647
|
0.419
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr1_+_201708940
|
0.411
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr11_-_67141008
|
0.408
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr9_-_79520878
|
0.405
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr6_+_117198375
|
0.390
|
NM_173560
|
RFX6
|
regulatory factor X, 6
|
|
chr2_-_175547471
|
0.390
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr2_-_175499275
|
0.389
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr4_+_174089889
|
0.385
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr8_+_1449531
|
0.383
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr21_-_16437125
|
0.380
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr15_-_56209305
|
0.378
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr5_-_38595506
|
0.374
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr5_-_38556682
|
0.374
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr11_+_131780537
|
0.370
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr11_+_118307072
|
0.367
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr2_+_30670082
|
0.366
|
NM_182551
NM_001002257
|
LCLAT1
|
lysocardiolipin acyltransferase 1
|
|
chr3_-_100712248
|
0.365
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr10_+_97889467
|
0.357
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr2_-_182545212
|
0.356
|
NM_002500
|
CERKL
NEUROD1
|
ceramide kinase-like
neurogenic differentiation 1
|
|
chr8_+_38585703
|
0.349
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr3_-_52713728
|
0.347
|
NM_018165
NM_181042
|
PBRM1
|
polybromo 1
|
|
chr8_-_82024277
|
0.342
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr9_-_27573444
|
0.341
|
NM_018325
|
C9orf72
|
chromosome 9 open reading frame 72
|
|
chr6_+_160390128
|
0.337
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr10_-_35363167
|
0.333
|
NM_001198778
NM_001198779
|
CUL2
|
cullin 2
|
|
chr17_+_26646120
|
0.332
|
NM_014573
|
TMEM97
|
transmembrane protein 97
|
|
chr2_-_217236666
|
0.331
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr4_+_160188997
|
0.331
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chrX_+_21857655
|
0.329
|
NM_015884
|
MBTPS2
|
membrane-bound transcription factor peptidase, site 2
|
|
chr4_-_139163222
|
0.329
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr5_+_154092461
|
0.328
|
NM_015315
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr2_-_145277879
|
0.326
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr8_+_98656329
|
0.326
|
NM_178812
|
MTDH
|
metadherin
|
|
chr10_+_26986557
|
0.325
|
NM_014317
|
PDSS1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1
|
|
chr6_-_52441815
|
0.322
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr18_-_65183922
|
0.322
|
NM_032160
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr2_-_166060552
|
0.320
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr12_-_90049818
|
0.319
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chrX_-_19688991
|
0.316
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_+_178310605
|
0.314
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr13_+_113623496
|
0.312
|
NM_001112732
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr12_-_54813010
|
0.311
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr8_+_38854431
|
0.310
|
NM_003816
|
ADAM9
|
ADAM metallopeptidase domain 9
|
|
chr5_+_134094460
|
0.306
|
NM_014829
|
DDX46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46
|
|
chr5_-_98262217
|
0.306
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr10_-_64028457
|
0.303
|
NM_145307
|
RTKN2
|
rhotekin 2
|
|
chr4_+_108745718
|
0.303
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr8_+_38644721
|
0.301
|
NM_001122824
NM_006283
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr12_-_82153092
|
0.294
|
NM_003625
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr17_+_74536114
|
0.294
|
NM_001077620
|
PRCD
|
progressive rod-cone degeneration
|
|
chr9_-_6645564
|
0.293
|
NM_000170
|
GLDC
|
glycine dehydrogenase (decarboxylating)
|
|
chr7_-_134001749
|
0.293
|
NM_032826
|
SLC35B4
|
solute carrier family 35, member B4
|
|
chr5_+_150827138
|
0.288
|
NM_078483
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr4_-_73434467
|
0.286
|
NM_014243
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3
|
|
chr20_+_48429249
|
0.286
|
NM_015266
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chr2_-_166650770
|
0.285
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr21_-_36421586
|
0.285
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr5_+_173315313
|
0.284
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr8_+_99129488
|
0.283
|
NM_001145860
NM_001145861
|
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae)
|
|
chr5_-_132299263
|
0.283
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr1_+_222791151
|
0.281
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr12_-_42538432
|
0.279
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr7_-_139477437
|
0.276
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr2_+_121554866
|
0.275
|
NM_005270
|
GLI2
|
GLI family zinc finger 2
|
|
chr2_+_191273012
|
0.275
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr3_-_101395611
|
0.274
|
NM_014415
|
ZBTB11
|
zinc finger and BTB domain containing 11
|
|
chr8_+_99130067
|
0.273
|
NM_015029
|
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae)
|
|
chr7_-_50800018
|
0.273
|
NM_001001549
NM_005311
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr12_-_112546562
|
0.271
|
NM_024953
|
NAA25
|
N(alpha)-acetyltransferase 25, NatB auxiliary subunit
|
|
chr15_-_56285732
|
0.270
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr5_+_70751435
|
0.267
|
NM_018429
|
BDP1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB
|
|
chr5_+_149109814
|
0.267
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr1_-_212004113
|
0.267
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr12_-_81991959
|
0.267
|
NM_001220477
NM_001220478
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr10_-_14372807
|
0.266
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr15_-_34630203
|
0.264
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr16_+_68298326
|
0.263
|
NM_001076785
NM_003983
|
SLC7A6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6
|
|
chrX_+_150151757
|
0.263
|
NM_005342
|
HMGB3
|
high mobility group box 3
|
|
chr2_-_153573858
|
0.262
|
NM_017892
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae)
|
|
chr9_-_14693459
|
0.260
|
NM_178566
|
ZDHHC21
|
zinc finger, DHHC-type containing 21
|
|
chr5_-_147162251
|
0.259
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr16_+_24740945
|
0.258
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr6_+_11538486
|
0.258
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr2_-_158731622
|
0.258
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr1_+_114496498
|
0.257
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr1_-_84464728
|
0.255
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr7_-_50772997
|
0.255
|
NM_001001550
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chrX_-_19817907
|
0.254
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr16_-_30022300
|
0.253
|
NM_003586
|
DOC2A
|
double C2-like domains, alpha
|
|
chr12_-_81763127
|
0.252
|
NM_001220479
NM_001220480
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr9_-_127703294
|
0.252
|
NM_002077
|
GOLGA1
|
golgin A1
|
|
chr6_-_7910716
|
0.251
|
NM_030810
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr14_-_52535945
|
0.250
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr10_-_35379223
|
0.250
|
NM_003591
|
CUL2
|
cullin 2
|
|
chr17_+_37617688
|
0.247
|
NM_015083
NM_016507
|
CDK12
|
cyclin-dependent kinase 12
|
|
chr8_+_37654395
|
0.247
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr12_-_77459198
|
0.247
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr6_+_158957467
|
0.245
|
NM_020823
|
TMEM181
|
transmembrane protein 181
|
|
chr12_-_82152269
|
0.245
|
NM_001220475
NM_001220476
NM_001220473
NM_001220474
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr4_+_108814419
|
0.245
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr17_+_76000248
|
0.244
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr3_+_132136503
|
0.244
|
NM_015268
|
DNAJC13
|
DnaJ (Hsp40) homolog, subfamily C, member 13
|
|
chr5_+_149151502
|
0.242
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chrX_-_40036581
|
0.241
|
NM_001123383
NM_001123384
|
BCOR
|
BCL6 corepressor
|
|
chr3_-_52719514
|
0.241
|
NM_018313
|
PBRM1
|
polybromo 1
|
|
chr12_-_104234968
|
0.241
|
NM_001031701
|
NT5DC3
|
5'-nucleotidase domain containing 3
|
|
chr4_-_52904424
|
0.241
|
NM_000232
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein)
|
|
chr15_+_96869156
|
0.241
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr12_-_46663207
|
0.240
|
NM_001077484
NM_030674
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr6_+_114178516
|
0.240
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr15_-_34610928
|
0.239
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr15_-_34629941
|
0.237
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr5_-_142783225
|
0.236
|
NM_000176
NM_001020825
NM_001024094
NM_001204258
NM_001204259
NM_001204260
NM_001204261
NM_001204262
NM_001204263
NM_001204264
NM_001204265
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr6_-_20212628
|
0.235
|
NM_001080480
|
MBOAT1
|
membrane bound O-acyltransferase domain containing 1
|
|
chr15_-_34629044
|
0.235
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_+_90287404
|
0.235
|
NM_018103
|
LRRC8D
|
leucine rich repeat containing 8 family, member D
|
|
chr13_+_113656027
|
0.233
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr5_+_175875355
|
0.232
|
NM_014613
|
FAF2
|
Fas associated factor family member 2
|
|
chr6_-_108395939
|
0.232
|
NM_014028
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chrX_+_17653412
|
0.231
|
NM_001136024
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr6_-_7910278
|
0.231
|
NM_001145549
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr10_-_35379480
|
0.231
|
NM_001198777
|
CUL2
|
cullin 2
|
|
chr2_-_158732341
|
0.231
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr1_-_86043932
|
0.230
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_+_173292306
|
0.230
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr16_-_2264776
|
0.230
|
NM_001042371
|
PGP
|
phosphoglycolate phosphatase
|
|
chr1_-_109506016
|
0.228
|
NM_001048210
NM_015127
|
CLCC1
AKNAD1
|
chloride channel CLIC-like 1
AKNA domain containing 1
|
|
chr19_+_11200037
|
0.227
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr12_+_52056547
|
0.226
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr9_-_136242969
|
0.226
|
NM_033161
|
SURF4
|
surfeit 4
|
|
chr2_-_214015053
|
0.226
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr2_-_214016332
|
0.225
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr12_+_51985016
|
0.225
|
NM_014191
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr1_+_41444968
|
0.224
|
NM_001905
|
CTPS
|
CTP synthase
|
|
chr16_-_1429612
|
0.223
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr16_+_53088791
|
0.221
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr20_+_39766128
|
0.220
|
NM_002660
NM_182811
|
PLCG1
|
phospholipase C, gamma 1
|
|
chr1_+_114493766
|
0.220
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chrX_+_17393537
|
0.220
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr15_+_99192696
|
0.218
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr14_+_52118539
|
0.218
|
NM_152330
|
FRMD6
|
FERM domain containing 6
|
|
chr15_+_96876568
|
0.217
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr3_+_238274
|
0.217
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr7_-_35077652
|
0.214
|
NM_015283
|
DPY19L1
|
dpy-19-like 1 (C. elegans)
|
|
chr9_-_107690526
|
0.214
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr19_+_37096202
|
0.213
|
NM_032825
|
ZNF382
|
zinc finger protein 382
|
|
chr14_+_51955854
|
0.212
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr7_-_2354050
|
0.212
|
NM_013321
|
SNX8
|
sorting nexin 8
|
|
chr16_+_71392615
|
0.211
|
NM_001740
NM_007088
|
CALB2
|
calbindin 2
|
|
chr18_+_54318561
|
0.210
|
NM_015285
NM_052834
|
WDR7
|
WD repeat domain 7
|
|
chr2_-_200322699
|
0.210
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|