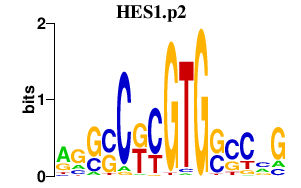
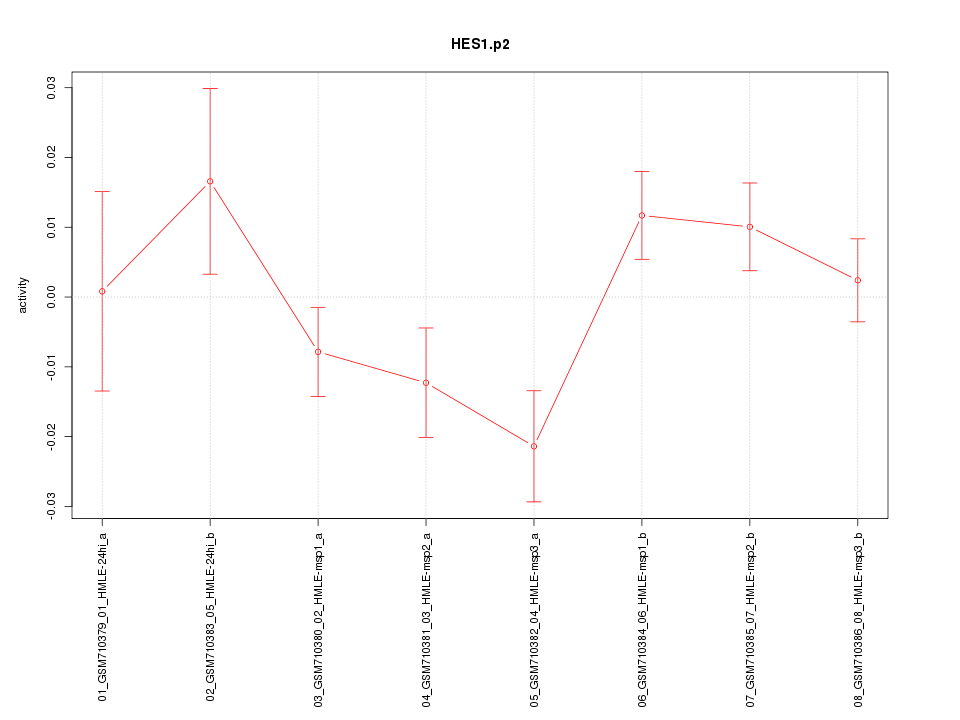
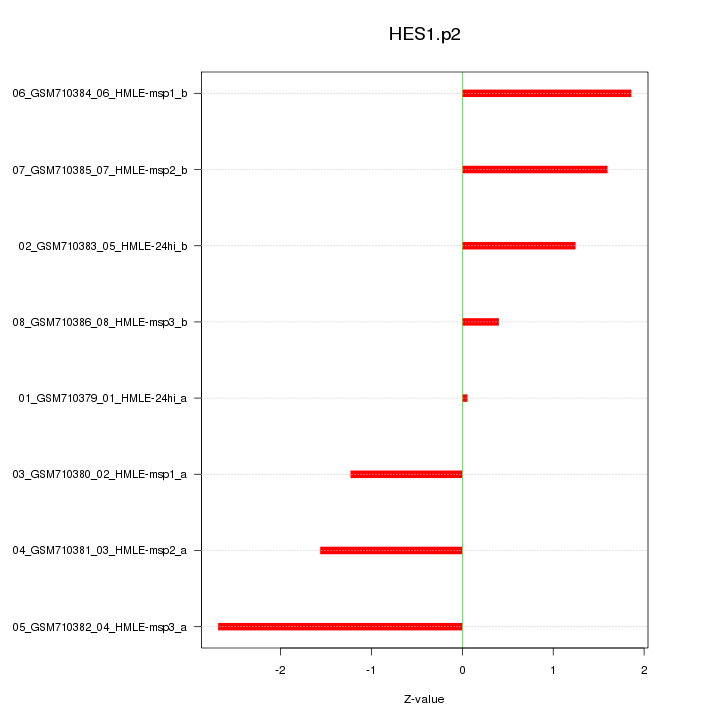
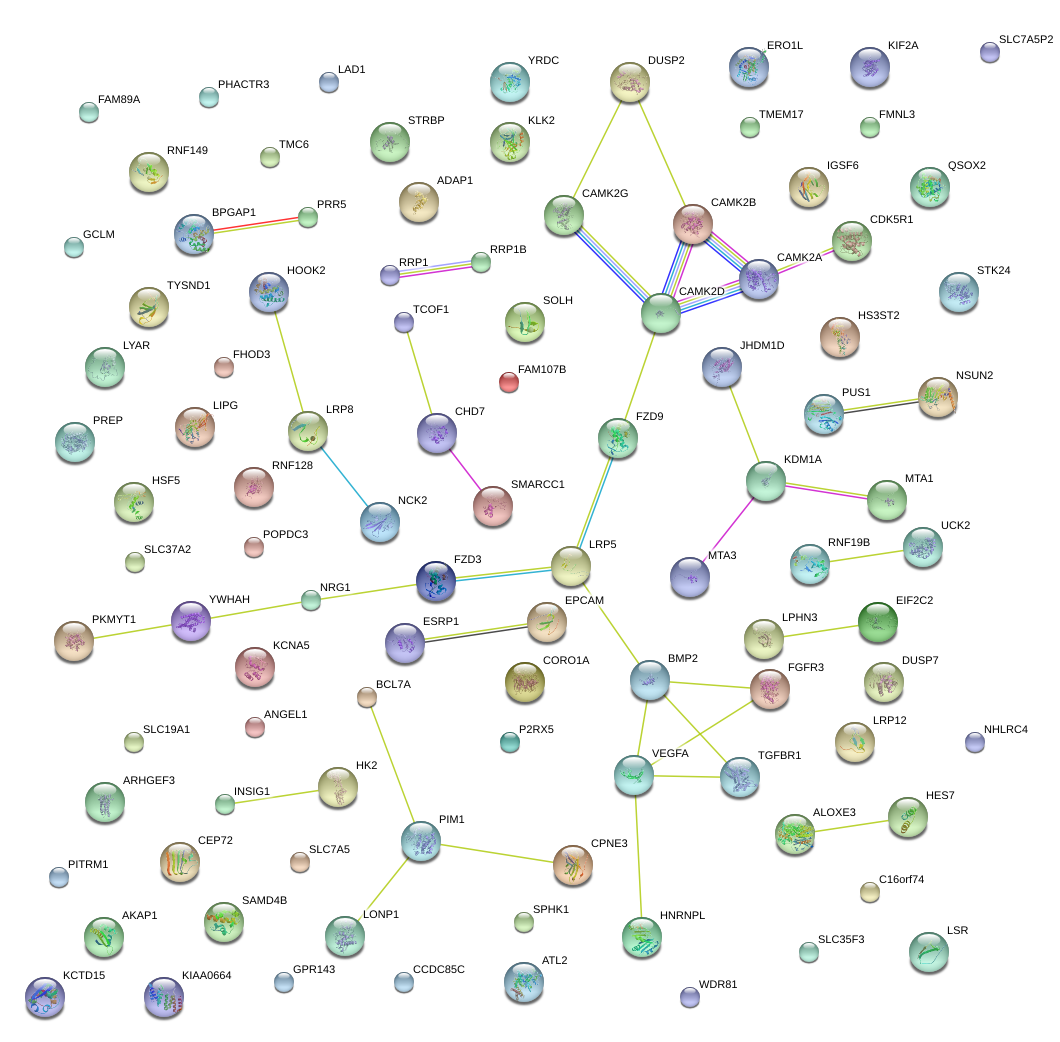
|
chr1_-_201368434
|
0.872
|
|
LAD1
|
ladinin 1
|
|
chr18_+_33877630
|
0.668
|
NM_025135
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr1_-_94374986
|
0.626
|
NM_002061
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr22_+_45098091
|
0.620
|
NM_181334
|
PRR5-ARHGAP8
PRR5
|
PRR5-ARHGAP8 readthrough
proline rich 5 (renal)
|
|
chr21_-_46962281
|
0.594
|
NM_001205206
NM_194255
|
SLC19A1
|
solute carrier family 19 (folate transporter), member 1
|
|
chr10_-_14646237
|
0.575
|
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr19_+_39833132
|
0.573
|
|
SAMD4B
|
sterile alpha motif domain containing 4B
|
|
chr1_+_234040547
|
0.568
|
NM_173508
|
SLC35F3
|
solute carrier family 35, member F3
|
|
chr2_-_96811138
|
0.555
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr1_-_33430285
|
0.544
|
NM_001127361
NM_153341
|
RNF19B
|
ring finger protein 19B
|
|
chr8_+_31497267
|
0.544
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr1_-_231175971
|
0.539
|
NM_198552
|
FAM89A
|
family with sequence similarity 89, member A
|
|
chr7_-_44365019
|
0.532
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr16_+_577833
|
0.526
|
NM_005632
|
SOLH
|
small optic lobes homolog (Drosophila)
|
|
chr5_+_149737296
|
0.498
|
|
TCOF1
|
Treacher Collins-Franceschetti syndrome 1
|
|
chr16_+_30194730
|
0.490
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr1_-_201368617
|
0.485
|
NM_005558
|
LAD1
|
ladinin 1
|
|
chr2_-_38604380
|
0.480
|
|
ATL2
|
atlastin GTPase 2
|
|
chr16_-_85784628
|
0.480
|
NM_206967
|
C16orf74
|
chromosome 16 open reading frame 74
|
|
chr9_-_126030792
|
0.477
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr2_-_38604430
|
0.477
|
NM_001135673
NM_022374
|
ATL2
|
atlastin GTPase 2
|
|
chr17_+_74380676
|
0.473
|
NM_001142601
NM_021972
NM_182965
|
SPHK1
|
sphingosine kinase 1
|
|
chr5_+_612404
|
0.471
|
NM_018140
|
CEP72
|
centrosomal protein 72kDa
|
|
chr19_+_1524072
|
0.467
|
NM_001243079
|
PLK5
|
polo-like kinase 5
|
|
chr5_+_149737201
|
0.456
|
NM_000356
NM_001008657
NM_001135243
NM_001135244
NM_001135245
NM_001195141
|
TCOF1
|
Treacher Collins-Franceschetti syndrome 1
|
|
chr6_-_105627857
|
0.454
|
NM_022361
|
POPDC3
|
popeye domain containing 3
|
|
chr5_+_612454
|
0.452
|
|
CEP72
|
centrosomal protein 72kDa
|
|
chr1_-_94375028
|
0.448
|
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr1_-_201368716
|
0.444
|
|
LAD1
|
ladinin 1
|
|
chr8_+_61591239
|
0.443
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr6_+_43738757
|
0.441
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr11_+_124933197
|
0.433
|
|
SLC37A2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr8_+_95653340
|
0.432
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr2_+_106361404
|
0.428
|
NM_001004722
NM_003581
|
NCK2
|
NCK adaptor protein 2
|
|
chr8_-_105601124
|
0.421
|
|
LRP12
|
low density lipoprotein receptor-related protein 12
|
|
chr1_-_94374949
|
0.418
|
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr7_+_26242007
|
0.418
|
|
|
|
|
chr16_-_87903028
|
0.417
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr8_+_95653399
|
0.414
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr16_+_22825806
|
0.412
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr1_-_38273852
|
0.409
|
NM_024640
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr1_+_15250578
|
0.409
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr19_+_35739838
|
0.402
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chrX_-_9733916
|
0.396
|
NM_000273
|
GPR143
|
G protein-coupled receptor 143
|
|
chr20_+_6748744
|
0.396
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr9_+_101867401
|
0.393
|
NM_001130916
NM_004612
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr17_-_76124767
|
0.392
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr7_+_155089614
|
0.375
|
|
INSIG1
|
insulin induced gene 1
|
|
chr14_-_77279242
|
0.369
|
NM_015305
|
ANGEL1
|
angel homolog 1 (Drosophila)
|
|
chr13_-_99229037
|
0.367
|
|
STK24
|
serine/threonine kinase 24
|
|
chr7_-_139876718
|
0.367
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr17_-_3599298
|
0.367
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr9_+_101867392
|
0.361
|
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr20_+_58152563
|
0.357
|
NM_001199505
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr8_+_28351694
|
0.356
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr4_-_4291817
|
0.353
|
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr9_-_139137646
|
0.352
|
NM_181701
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2
|
|
chr2_-_62733475
|
0.352
|
NM_198276
|
TMEM17
|
transmembrane protein 17
|
|
chr14_-_53162418
|
0.350
|
NM_014584
|
ERO1L
|
ERO1-like (S. cerevisiae)
|
|
chr19_+_34287662
|
0.349
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr10_-_71905947
|
0.348
|
|
TYSND1
|
trypsin domain containing 1
|
|
chr9_-_126030854
|
0.348
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr1_+_23345985
|
0.344
|
|
KDM1A
|
lysine (K)-specific demethylase 1A
|
|
chr8_+_61591358
|
0.344
|
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr17_-_3599359
|
0.343
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chrX_+_105969875
|
0.342
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr2_+_47596286
|
0.341
|
NM_002354
|
EPCAM
|
epithelial cell adhesion molecule
|
|
chr8_-_141645603
|
0.339
|
NM_001164623
NM_012154
|
EIF2C2
|
eukaryotic translation initiation factor 2C, 2
|
|
chr17_+_55162530
|
0.337
|
NM_003488
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr1_-_38273846
|
0.334
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr7_-_994259
|
0.333
|
NM_006869
|
ADAP1
|
ArfGAP with dual PH domains 1
|
|
chr4_+_1795004
|
0.331
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr2_-_75937707
|
0.330
|
NM_001201334
|
GCFC2
|
GC-rich sequence DNA-binding factor 2
|
|
chr18_+_47088400
|
0.327
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr1_-_53793583
|
0.326
|
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr4_-_4291889
|
0.324
|
NM_017816
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr17_+_30813928
|
0.321
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr1_+_23345928
|
0.318
|
NM_001009999
NM_015013
|
KDM1A
|
lysine (K)-specific demethylase 1A
|
|
chr11_+_124933007
|
0.316
|
NM_001145290
NM_198277
|
SLC37A2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr17_+_1633754
|
0.315
|
|
WDR81
|
WD repeat domain 81
|
|
chr3_-_52090090
|
0.314
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr18_-_77793914
|
0.313
|
|
|
|
|
chr14_+_105886146
|
0.312
|
NM_001203258
NM_004689
|
MTA1
|
metastasis associated 1
|
|
chr12_+_122459791
|
0.311
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr5_-_6633149
|
0.311
|
|
NSUN2
|
NOP2/Sun domain family, member 2
|
|
chr17_-_56565700
|
0.311
|
NM_001080439
|
HSF5
|
heat shock transcription factor family member 5
|
|
chr19_+_35739778
|
0.309
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr1_+_165797060
|
0.308
|
|
UCK2
|
uridine-cytidine kinase 2
|
|
chr1_-_53793743
|
0.306
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr2_-_101925051
|
0.293
|
NM_173647
|
RNF149
|
ring finger protein 149
|
|
chr3_-_57113292
|
0.293
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr6_+_37137882
|
0.292
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr17_+_74380943
|
0.292
|
|
SPHK1
|
sphingosine kinase 1
|
|
chr21_+_45209398
|
0.291
|
NM_003683
|
RRP1
|
ribosomal RNA processing 1 homolog (S. cerevisiae)
|
|
chr2_+_75061234
|
0.290
|
|
HK2
|
hexokinase 2
|
|
chr17_-_8022134
|
0.290
|
NM_001165960
|
ALOXE3
|
arachidonate lipoxygenase 3
|
|
chr17_-_2614926
|
0.287
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr11_+_68079999
|
0.285
|
NM_002335
|
LRP5
|
low density lipoprotein receptor-related protein 5
|
|
chr22_+_32340435
|
0.285
|
NM_003405
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr8_+_87526666
|
0.284
|
|
CPNE3
|
copine III
|
|
chr19_-_39340362
|
0.282
|
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L
|
|
chr12_+_132413776
|
0.277
|
NM_001002019
NM_025215
|
PUS1
|
pseudouridylate synthase 1
|
|
chr16_+_610393
|
0.276
|
NM_145270
|
C16orf11
|
chromosome 16 open reading frame 11
|
|
chr6_-_105850765
|
0.275
|
|
PREP
|
prolyl endopeptidase
|
|
chr16_-_3030391
|
0.272
|
|
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1
|
|
chr3_-_47823419
|
0.272
|
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr19_+_35739519
|
0.272
|
NM_015925
NM_205834
NM_205835
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr14_-_100070442
|
0.272
|
NM_001144995
|
CCDC85C
|
coiled-coil domain containing 85C
|
|
chr8_+_87526646
|
0.270
|
NM_003909
|
CPNE3
|
copine III
|
|
chr17_-_8027396
|
0.268
|
NM_001165967
NM_032580
|
HES7
|
hairy and enhancer of split 7 (Drosophila)
|
|
chr18_-_12377168
|
0.268
|
|
AFG3L2
|
AFG3 ATPase family gene 3-like 2 (S. cerevisiae)
|
|
chr13_-_99229117
|
0.267
|
|
STK24
|
serine/threonine kinase 24
|
|
chr14_-_53162198
|
0.266
|
|
ERO1L
|
ERO1-like (S. cerevisiae)
|
|
chr16_-_3030473
|
0.265
|
NM_004203
NM_182687
|
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1
|
|
chr15_-_26108236
|
0.265
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr4_+_57774365
|
0.264
|
|
REST
|
RE1-silencing transcription factor
|
|
chr2_+_131674223
|
0.263
|
NM_015320
NM_032995
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4
|
|
chr1_-_175161832
|
0.262
|
NM_001162895
NM_014656
NM_001162893
NM_001162894
|
KIAA0040
|
KIAA0040
|
|
chr7_-_98030165
|
0.261
|
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
chr8_-_105601234
|
0.260
|
NM_001135703
NM_013437
|
LRP12
|
low density lipoprotein receptor-related protein 12
|
|
chr16_+_22825481
|
0.259
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr17_+_74381186
|
0.259
|
NM_001142602
|
SPHK1
|
sphingosine kinase 1
|
|
chr14_-_23834417
|
0.258
|
|
EFS
|
embryonal Fyn-associated substrate
|
|
chr19_+_4402658
|
0.256
|
NM_005483
|
CHAF1A
|
chromatin assembly factor 1, subunit A (p150)
|
|
chr1_-_17764811
|
0.256
|
|
RCC2
|
regulator of chromosome condensation 2
|
|
chr17_+_45727303
|
0.255
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr1_-_85462622
|
0.255
|
NM_153259
|
MCOLN2
|
mucolipin 2
|
|
chr17_-_3599487
|
0.253
|
NM_001204519
NM_001204520
NM_002561
NM_175080
|
P2RX5-TAX1BP3
P2RX5
|
P2RX5-TAX1BP3 readthrough
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr3_-_10547221
|
0.251
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr21_+_35445808
|
0.250
|
NM_032476
NM_006933
|
MRPS6
SLC5A3
|
mitochondrial ribosomal protein S6
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3
|
|
chr3_+_57994087
|
0.249
|
NM_001164317
NM_001164318
NM_001164319
NM_001457
|
FLNB
|
filamin B, beta
|
|
chr19_+_39833099
|
0.249
|
NM_018028
|
SAMD4B
|
sterile alpha motif domain containing 4B
|
|
chr16_-_4401265
|
0.249
|
NM_016069
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae)
|
|
chr7_+_24612714
|
0.247
|
NM_016447
|
MPP6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6)
|
|
chr1_+_46713385
|
0.245
|
|
RAD54L
|
RAD54-like (S. cerevisiae)
|
|
chr12_+_8850479
|
0.244
|
NM_020734
|
RIMKLB
|
ribosomal modification protein rimK-like family member B
|
|
chr17_+_45727274
|
0.243
|
NM_002265
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr1_-_11120081
|
0.243
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr13_-_20806533
|
0.242
|
NM_001110219
NM_001110220
NM_001110221
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr2_-_216878314
|
0.241
|
NM_018000
|
MREG
|
melanoregulin
|
|
chr16_-_68269948
|
0.240
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chrX_-_7066146
|
0.238
|
NM_001135565
NM_001178135
NM_001178136
NM_012080
|
HDHD1
|
haloacid dehalogenase-like hydrolase domain containing 1
|
|
chr6_-_143832770
|
0.238
|
NM_032020
|
FUCA2
|
fucosidase, alpha-L- 2, plasma
|
|
chr1_-_6662681
|
0.238
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr13_-_99229226
|
0.238
|
NM_001032296
|
STK24
|
serine/threonine kinase 24
|
|
chr1_+_229761988
|
0.237
|
|
URB2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae)
|
|
chr11_+_65029312
|
0.237
|
NM_002689
|
POLA2
|
polymerase (DNA directed), alpha 2 (70kD subunit)
|
|
chr16_-_1968230
|
0.236
|
NM_001009606
|
HS3ST6
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 6
|
|
chr22_+_29876180
|
0.236
|
NM_021076
|
NEFH
|
neurofilament, heavy polypeptide
|
|
chr10_+_134210691
|
0.234
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr7_+_116312412
|
0.233
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr14_+_103243832
|
0.233
|
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr2_-_70781146
|
0.232
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr14_-_53162370
|
0.232
|
|
ERO1L
|
ERO1-like (S. cerevisiae)
|
|
chr7_-_994008
|
0.232
|
|
ADAP1
|
ArfGAP with dual PH domains 1
|
|
chr17_-_17109601
|
0.230
|
|
PLD6
|
phospholipase D family, member 6
|
|
chr22_+_25960885
|
0.230
|
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr7_-_45151272
|
0.230
|
|
TBRG4
|
transforming growth factor beta regulator 4
|
|
chr8_-_145515014
|
0.228
|
|
BOP1
|
block of proliferation 1
|
|
chr8_-_145515099
|
0.227
|
NM_015201
|
BOP1
|
block of proliferation 1
|
|
chr6_-_105850958
|
0.227
|
|
PREP
|
prolyl endopeptidase
|
|
chr8_-_145515089
|
0.226
|
|
BOP1
|
block of proliferation 1
|
|
chr6_-_105850976
|
0.226
|
NM_002726
|
PREP
|
prolyl endopeptidase
|
|
chr19_-_1863425
|
0.226
|
|
KLF16
|
Kruppel-like factor 16
|
|
chr14_+_103243813
|
0.225
|
NM_001199427
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr17_+_45726838
|
0.225
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr8_-_125740378
|
0.225
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr9_+_133971862
|
0.224
|
NM_001185095
NM_001185096
NM_031426
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr17_+_29718777
|
0.223
|
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr8_-_145515071
|
0.222
|
|
BOP1
|
block of proliferation 1
|
|
chr3_+_184032319
|
0.222
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr11_+_93862143
|
0.221
|
|
PANX1
|
pannexin 1
|
|
chr19_+_38397841
|
0.221
|
NM_015073
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3
|
|
chr4_+_1873035
|
0.221
|
NM_001042424
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr1_+_117452359
|
0.219
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr17_-_41623246
|
0.219
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr17_+_29718641
|
0.219
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr17_-_8021806
|
0.215
|
NM_021628
|
ALOXE3
|
arachidonate lipoxygenase 3
|
|
chr1_-_229761793
|
0.215
|
NM_001025247
NM_014409
|
TAF5L
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa
|
|
chr19_-_46272102
|
0.215
|
|
|
|
|
chr1_-_220101925
|
0.214
|
NM_018713
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr11_-_119234628
|
0.214
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr17_-_40428311
|
0.214
|
|
STAT5B
|
signal transducer and activator of transcription 5B
|
|
chr1_+_118472364
|
0.214
|
NM_006784
|
WDR3
|
WD repeat domain 3
|
|
chrX_-_153285262
|
0.213
|
|
IRAK1
|
interleukin-1 receptor-associated kinase 1
|
|
chr6_+_43738394
|
0.213
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr3_-_189838673
|
0.212
|
|
LEPREL1
|
leprecan-like 1
|
|
chr19_+_1248548
|
0.212
|
NM_177401
|
MIDN
|
midnolin
|
|
chr9_-_23821842
|
0.212
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr9_-_23821808
|
0.211
|
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr5_+_90676131
|
0.211
|
|
LOC100129716
|
uncharacterized LOC100129716
|
|
chr17_-_76124791
|
0.211
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr6_+_43738177
|
0.211
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr5_-_1494872
|
0.211
|
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1
|
|
chr16_+_30194925
|
0.210
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr19_+_984316
|
0.209
|
NM_024100
|
WDR18
|
WD repeat domain 18
|
|
chr13_-_77460432
|
0.208
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr7_-_98030426
|
0.208
|
NM_018842
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
chrX_+_38420690
|
0.207
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr3_+_184032414
|
0.206
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|