|
chr14_-_105634708
|
4.149
|
|
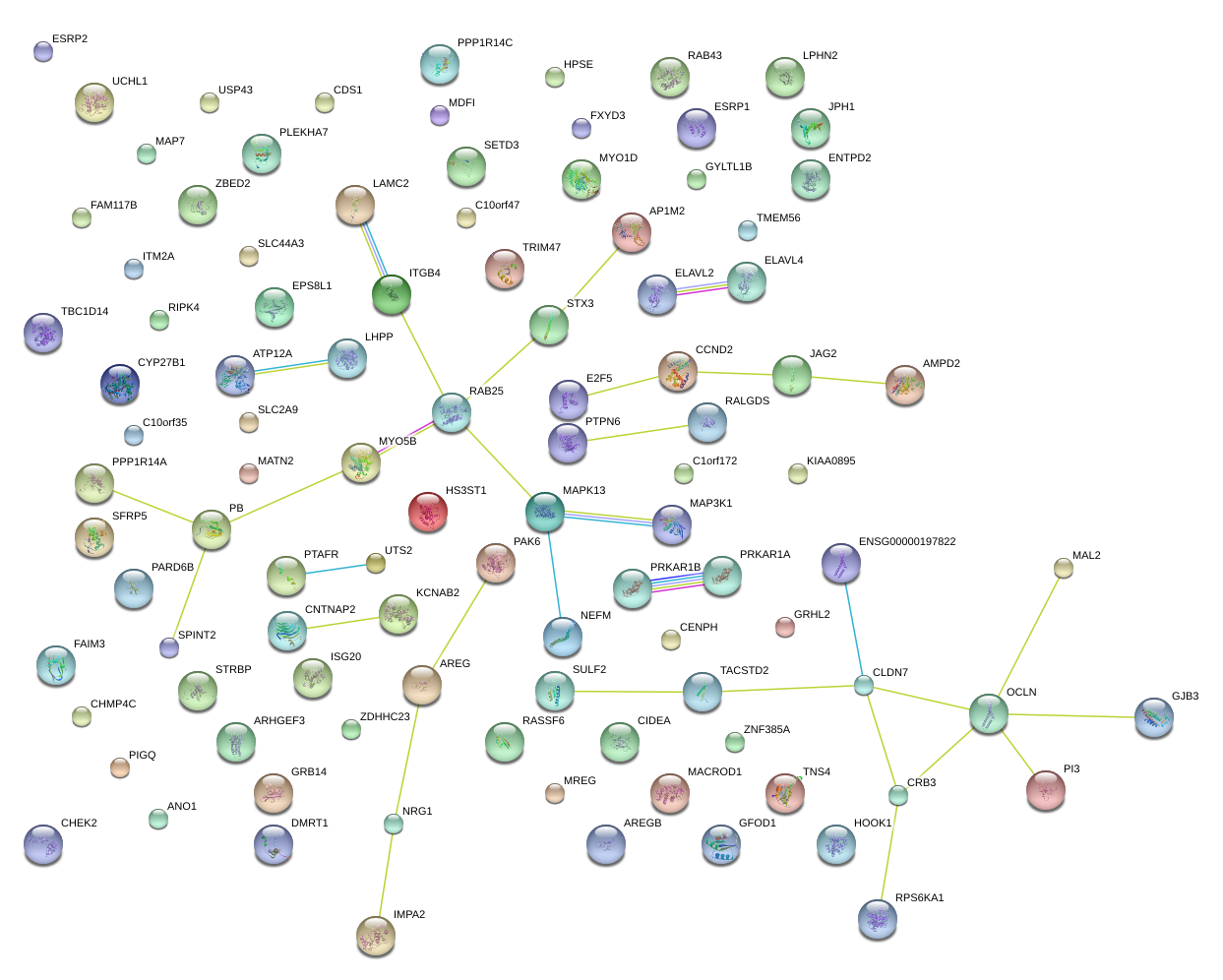
JAG2
|
jagged 2
|
|
chr8_+_95653272
|
3.164
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr1_-_27286875
|
2.846
|
NM_152365
|
C1orf172
|
chromosome 1 open reading frame 172
|
|
chr8_-_75233461
|
2.649
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr1_+_156030965
|
2.238
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr11_+_69924407
|
2.193
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr19_+_35607218
|
2.176
|
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr14_-_105635114
|
1.998
|
NM_002226
NM_145159
|
JAG2
|
jagged 2
|
|
chr3_-_111314181
|
1.863
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr19_-_10697907
|
1.848
|
NM_005498
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit
|
|
chr19_+_38755097
|
1.848
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_38755238
|
1.825
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr8_+_31497267
|
1.774
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr6_-_136871956
|
1.758
|
NM_001198616
NM_001198617
NM_003980
|
MAP7
|
microtubule-associated protein 7
|
|
chr19_+_38755422
|
1.754
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr12_+_4382882
|
1.730
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr9_-_136024522
|
1.692
|
NM_001042368
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr7_+_145813828
|
1.661
|
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr17_-_7166263
|
1.504
|
NM_001185023
NM_001307
|
CLDN7
|
claudin 7
|
|
chr9_-_23821477
|
1.475
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr1_+_183155393
|
1.442
|
|
LAMC2
|
laminin, gamma 2
|
|
chr1_+_35247838
|
1.399
|
NM_001005752
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr1_+_60280462
|
1.383
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr10_+_71390002
|
1.361
|
NM_145306
|
C10orf35
|
chromosome 10 open reading frame 35
|
|
chr6_-_136871542
|
1.345
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr5_+_68788387
|
1.326
|
NM_001205254
|
OCLN
|
occludin
|
|
chr8_+_102504836
|
1.305
|
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr8_+_102504661
|
1.274
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr8_+_82644663
|
1.258
|
NM_152284
|
CHMP4C
|
charged multivesicular body protein 4C
|
|
chr16_-_68269948
|
1.252
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr3_-_57113292
|
1.249
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr12_-_58160823
|
1.209
|
NM_000785
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr17_-_31203889
|
1.196
|
NM_015194
|
MYO1D
|
myosin ID
|
|
chr2_+_203499819
|
1.114
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr12_-_54778456
|
1.105
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr8_+_120220554
|
1.075
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr13_+_25254548
|
1.051
|
NM_001185085
NM_001676
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide
|
|
chr17_+_9548853
|
1.035
|
NM_153210
|
USP43
|
ubiquitin specific peptidase 43
|
|
chr1_+_26856259
|
1.009
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr4_-_11430502
|
0.991
|
NM_005114
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1
|
|
chr1_+_82266081
|
0.969
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr9_-_125941304
|
0.967
|
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr1_-_207095198
|
0.959
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr9_-_139948448
|
0.959
|
NM_001246
NM_203468
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
|
chr6_+_150464156
|
0.954
|
NM_030949
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C
|
|
chr6_+_41606193
|
0.951
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr1_-_59042827
|
0.941
|
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr2_-_165478357
|
0.936
|
NM_004490
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr17_-_7165788
|
0.931
|
|
CLDN7
|
claudin 7
|
|
chr1_+_95558072
|
0.924
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr18_-_47721165
|
0.917
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr11_+_45944155
|
0.904
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr8_+_86089618
|
0.889
|
NM_001083588
NM_001951
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr19_+_55587220
|
0.887
|
NM_133180
|
EPS8L1
|
EPS8-like 1
|
|
chr20_+_43803539
|
0.879
|
NM_002638
|
PI3
|
peptidase inhibitor 3, skin-derived
|
|
chr12_+_7060543
|
0.842
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr12_+_7060531
|
0.836
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr4_-_74486100
|
0.835
|
NM_177532
NM_201431
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chr2_-_216878314
|
0.831
|
NM_018000
|
MREG
|
melanoregulin
|
|
chrX_-_78622711
|
0.830
|
|
ITM2A
|
integral membrane protein 2A
|
|
chr1_-_7973286
|
0.820
|
|
UTS2
|
urotensin 2
|
|
chr12_+_7060517
|
0.813
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr1_-_28503454
|
0.797
|
NM_000952
NM_001164723
|
PTAFR
|
platelet-activating factor receptor
|
|
chr12_+_7060538
|
0.791
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr18_+_11981552
|
0.789
|
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr4_-_10023094
|
0.782
|
NM_020041
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9
|
|
chr18_+_11981421
|
0.778
|
NM_014214
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr17_-_31204071
|
0.777
|
|
MYO1D
|
myosin ID
|
|
chr8_+_32405727
|
0.754
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr12_+_7060554
|
0.754
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr11_-_63933493
|
0.752
|
NM_014067
|
MACROD1
|
MACRO domain containing 1
|
|
chr8_+_86089641
|
0.743
|
|
|
|
|
chr1_+_95582827
|
0.739
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr9_+_841689
|
0.736
|
NM_021951
|
DMRT1
|
doublesex and mab-3 related transcription factor 1
|
|
chr19_+_6464259
|
0.735
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr17_+_73717596
|
0.722
|
|
ITGB4
|
integrin, beta 4
|
|
chr17_+_73717556
|
0.719
|
|
ITGB4
|
integrin, beta 4
|
|
chr10_+_11865310
|
0.711
|
NM_153256
|
C10orf47
|
chromosome 10 open reading frame 47
|
|
chr1_+_26856252
|
0.707
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr7_-_36406446
|
0.704
|
NM_001199707
NM_001199706
NM_015314
NM_001199708
|
KIAA0895
|
KIAA0895
|
|
chr6_+_36098261
|
0.703
|
NM_002754
|
MAPK13
|
mitogen-activated protein kinase 13
|
|
chr17_-_38657814
|
0.699
|
NM_032865
|
TNS4
|
tensin 4
|
|
chr12_+_7060433
|
0.698
|
NM_002831
NM_080549
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr8_+_98881277
|
0.693
|
NM_002380
NM_030583
|
MATN2
|
matrilin 2
|
|
chr7_-_752742
|
0.689
|
NM_001164761
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta
|
|
chr4_+_6911456
|
0.688
|
NM_020773
|
TBC1D14
|
TBC1 domain family, member 14
|
|
chr21_-_43187265
|
0.684
|
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr8_+_24772454
|
0.678
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr1_+_95285900
|
0.678
|
NM_001114106
|
SLC44A3
|
solute carrier family 44, member 3
|
|
chr21_-_43187197
|
0.677
|
NM_020639
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr4_+_85504056
|
0.677
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr20_-_46414636
|
0.675
|
NM_018837
|
SULF2
|
sulfatase 2
|
|
chr18_+_12254317
|
0.674
|
NM_001279
|
CIDEA
|
cell death-inducing DFFA-like effector a
|
|
chr4_+_75310852
|
0.670
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr14_-_99947080
|
0.668
|
|
SETD3
|
SET domain containing 3
|
|
chr1_+_6052758
|
0.664
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr1_+_6052699
|
0.663
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr11_+_59522801
|
0.662
|
|
STX3
|
syntaxin 3
|
|
chr11_-_17035948
|
0.658
|
NM_175058
|
PLEKHA7
|
pleckstrin homology domain containing, family A member 7
|
|
chr17_+_73717515
|
0.657
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr20_+_49348050
|
0.652
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr11_+_59522905
|
0.650
|
|
STX3
|
syntaxin 3
|
|
chr17_-_73874634
|
0.644
|
NM_033452
|
TRIM47
|
tripartite motif containing 47
|
|
chr15_+_89182038
|
0.643
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr15_+_40509628
|
0.643
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr6_-_13486986
|
0.643
|
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr19_-_38746978
|
0.640
|
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A
|
|
chr9_+_22447064
|
0.638
|
|
|
|
|
chr10_-_99531712
|
0.636
|
NM_003015
|
SFRP5
|
secreted frizzled-related protein 5
|
|
chr4_+_6911094
|
0.631
|
NM_001113361
|
TBC1D14
|
TBC1 domain family, member 14
|
|
chr5_+_56111379
|
0.629
|
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr8_+_32504250
|
0.629
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr3_+_113666544
|
0.626
|
NM_173570
|
ZDHHC23
|
zinc finger, DHHC-type containing 23
|
|
chr11_-_63933204
|
0.624
|
|
MACROD1
|
MACRO domain containing 1
|
|
chr10_+_126150340
|
0.623
|
NM_001167880
NM_022126
|
LHPP
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase
|
|
chr4_+_41258941
|
0.620
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr1_+_110163667
|
0.618
|
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr5_+_68485367
|
0.616
|
NM_022909
|
CENPH
|
centromere protein H
|
|
chr3_-_128840343
|
0.605
|
|
RAB43
|
RAB43, member RAS oncogene family
|
|
chr4_-_84256304
|
0.604
|
NM_001166498
NM_006665
|
HPSE
|
heparanase
|
|
chr1_-_32801606
|
0.602
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr3_-_128840605
|
0.600
|
NM_001204886
NM_001204887
NM_001204888
NM_198490
NM_001204885
|
RAB43
|
RAB43, member RAS oncogene family
|
|
chr17_+_40714420
|
0.599
|
|
|
|
|
chr2_-_165477818
|
0.594
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr5_+_68710938
|
0.594
|
NM_001038603
NM_001244734
|
MARVELD2
|
MARVEL domain containing 2
|
|
chr6_+_80340971
|
0.592
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr6_-_2245655
|
0.592
|
NM_001500
|
GMDS
|
GDP-mannose 4,6-dehydratase
|
|
chr1_-_160068261
|
0.588
|
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr7_-_16844606
|
0.585
|
NM_006408
|
AGR2
|
anterior gradient 2 homolog (Xenopus laevis)
|
|
chr14_+_65171149
|
0.583
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr15_+_41136178
|
0.583
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr1_+_206730456
|
0.579
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr12_+_104850762
|
0.579
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr4_+_6911537
|
0.578
|
|
TBC1D14
|
TBC1 domain family, member 14
|
|
chr12_+_122356446
|
0.571
|
NM_001178003
NM_144668
|
WDR66
|
WD repeat domain 66
|
|
chr20_+_30639981
|
0.569
|
NM_001172129
NM_001172130
NM_001172131
NM_001172132
NM_001172133
NM_002110
|
HCK
|
hemopoietic cell kinase
|
|
chr7_-_37488554
|
0.551
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr1_-_160068617
|
0.545
|
NM_001206665
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr16_-_48269087
|
0.542
|
NM_032583
NM_145186
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11
|
|
chr17_-_73874056
|
0.541
|
|
TRIM47
|
tripartite motif containing 47
|
|
chrX_+_37545008
|
0.541
|
NM_021083
|
XK
|
X-linked Kx blood group (McLeod syndrome)
|
|
chr6_-_31550065
|
0.539
|
NM_002341
NM_009588
|
LTB
|
lymphotoxin beta (TNF superfamily, member 3)
|
|
chrX_-_78623006
|
0.538
|
NM_001171581
NM_004867
|
ITM2A
|
integral membrane protein 2A
|
|
chr11_+_68079999
|
0.530
|
NM_002335
|
LRP5
|
low density lipoprotein receptor-related protein 5
|
|
chr14_-_23623493
|
0.529
|
NM_182728
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr19_+_45843997
|
0.525
|
NM_177417
|
KLC3
|
kinesin light chain 3
|
|
chr1_-_160068359
|
0.516
|
NM_052868
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr8_-_24772229
|
0.513
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr2_-_165477986
|
0.510
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr9_+_135854097
|
0.504
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr4_-_90758126
|
0.497
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr17_-_74533623
|
0.497
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr18_+_55711890
|
0.493
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr11_+_67070918
|
0.492
|
NM_017857
|
SSH3
|
slingshot homolog 3 (Drosophila)
|
|
chr6_-_32157511
|
0.490
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chr1_+_15256295
|
0.490
|
NM_001018001
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr11_-_19223516
|
0.489
|
NM_001127656
NM_003476
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein)
|
|
chr1_+_183155173
|
0.484
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr7_-_41742666
|
0.481
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr3_+_50192420
|
0.481
|
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr11_+_67070987
|
0.480
|
|
SSH3
|
slingshot homolog 3 (Drosophila)
|
|
chr21_+_43710190
|
0.479
|
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr4_+_3465032
|
0.478
|
NM_001164673
NM_173660
|
DOK7
|
docking protein 7
|
|
chr7_-_156803224
|
0.472
|
NM_005515
|
MNX1
|
motor neuron and pancreas homeobox 1
|
|
chr6_+_33043702
|
0.460
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr1_-_217311095
|
0.459
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_+_3371020
|
0.458
|
NM_014448
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr5_+_56110899
|
0.458
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr20_-_6103860
|
0.457
|
|
FERMT1
|
fermitin family member 1
|
|
chr12_+_104850641
|
0.455
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr14_-_51561657
|
0.455
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr1_+_6106173
|
0.454
|
NM_001199863
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr2_-_220436267
|
0.453
|
NM_001173408
NM_001173431
NM_015311
|
OBSL1
|
obscurin-like 1
|
|
chr17_-_8151353
|
0.452
|
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr9_+_22446783
|
0.451
|
NM_022160
|
DMRTA1
|
DMRT-like family A1
|
|
chr1_-_6557448
|
0.450
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr19_-_38747171
|
0.450
|
NM_001243947
NM_033256
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A
|
|
chr9_-_135996287
|
0.448
|
NM_006266
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr7_+_26191732
|
0.443
|
NM_004289
|
NFE2L3
|
nuclear factor (erythroid-derived 2)-like 3
|
|
chr7_+_156803599
|
0.439
|
|
LOC645249
|
uncharacterized LOC645249
|
|
chr8_-_66753968
|
0.438
|
NM_001242318
|
PDE7A
|
phosphodiesterase 7A
|
|
chr20_-_6103562
|
0.437
|
|
FERMT1
|
fermitin family member 1
|
|
chr1_-_160990862
|
0.436
|
|
F11R
|
F11 receptor
|
|
chr1_+_6052357
|
0.435
|
NM_001199861
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr16_+_84682126
|
0.433
|
NM_024731
|
KLHL36
|
kelch-like 36 (Drosophila)
|
|
chr4_-_110650965
|
0.433
|
NM_030821
|
PLA2G12A
|
phospholipase A2, group XIIA
|
|
chr19_-_41220977
|
0.431
|
|
ADCK4
|
aarF domain containing kinase 4
|
|
chr1_+_183155361
|
0.430
|
|
LAMC2
|
laminin, gamma 2
|
|
chr17_-_9694613
|
0.429
|
NM_001105571
NM_001220493
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chr4_+_81106423
|
0.426
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr1_+_206730497
|
0.426
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr9_-_131940481
|
0.425
|
|
IER5L
|
immediate early response 5-like
|
|
chr1_-_109940497
|
0.424
|
NM_002959
|
SORT1
|
sortilin 1
|
|
chr12_+_110152186
|
0.423
|
NM_032829
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr6_-_40555111
|
0.421
|
NM_020737
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2
|
|
chr4_+_41258895
|
0.421
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr20_+_45523523
|
0.418
|
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr1_+_226411318
|
0.416
|
NM_031944
|
MIXL1
|
Mix paired-like homeobox
|
|
chr3_+_10857867
|
0.415
|
NM_014229
|
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr11_-_105948464
|
0.414
|
NM_152433
NM_198439
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3
|