|
chr14_-_60337350
|
5.734
|
NM_021136
|
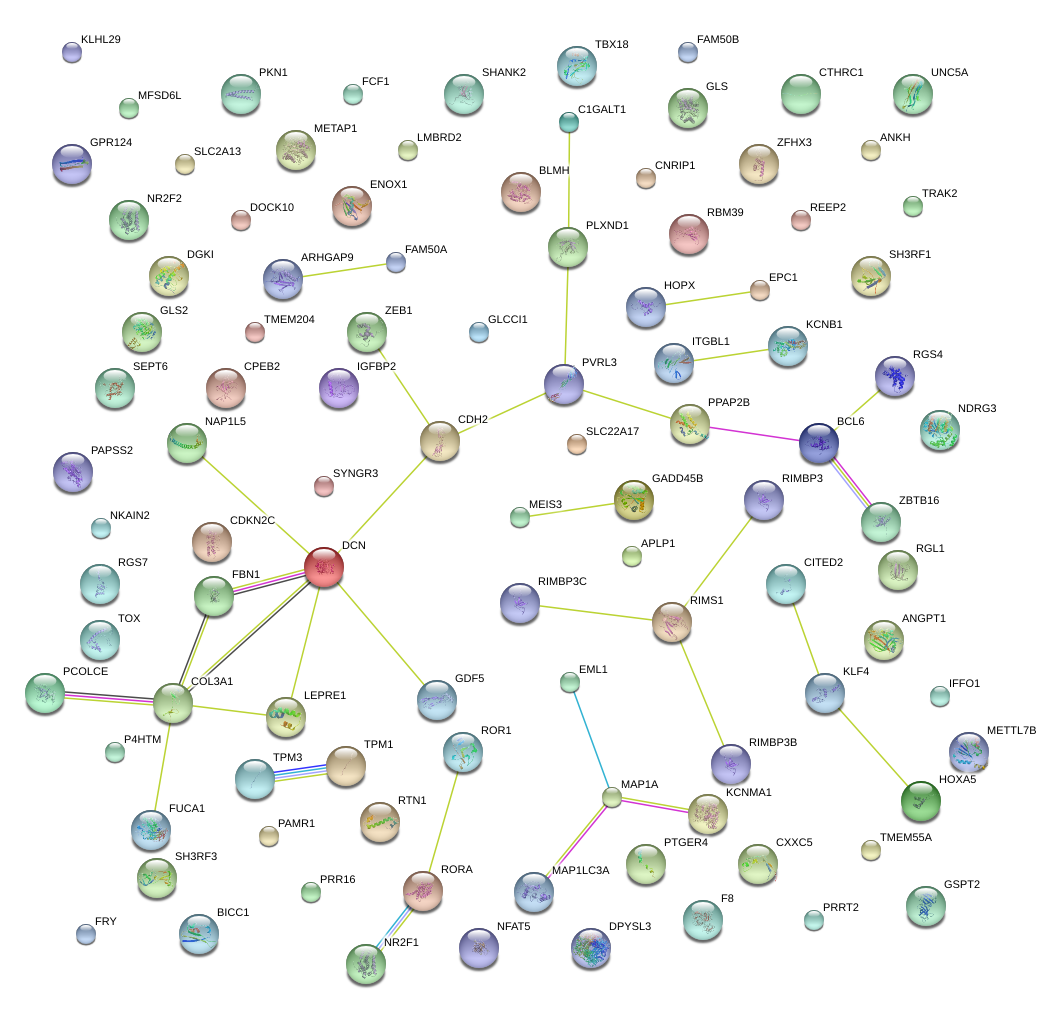
RTN1
|
reticulon 1
|
|
chr5_+_92920592
|
4.031
|
|
NR2F1
NR2F2
|
nuclear receptor subfamily 2, group F, member 1
nuclear receptor subfamily 2, group F, member 2
|
|
chr15_-_48937068
|
3.898
|
|
FBN1
|
fibrillin 1
|
|
chr3_+_110790460
|
2.657
|
NM_001243286
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr6_+_72596699
|
2.625
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_-_57045214
|
2.614
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr13_+_102104941
|
2.362
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr4_+_15004297
|
2.285
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr8_+_37654757
|
2.210
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr7_-_137531585
|
2.191
|
NM_004717
|
DGKI
|
diacylglycerol kinase, iota
|
|
chr8_+_37654406
|
2.158
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr1_-_57044980
|
2.143
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr12_+_90102639
|
2.115
|
|
LOC338758
|
uncharacterized LOC338758
|
|
chr1_-_57045235
|
2.111
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr10_-_79397394
|
2.038
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr11_-_35547144
|
2.006
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr5_+_95067511
|
1.979
|
|
|
|
|
chr10_-_79397204
|
1.935
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr18_-_25756946
|
1.918
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr2_-_225907312
|
1.885
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr8_+_37654395
|
1.808
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chrX_+_153672490
|
1.795
|
|
FAM50A
|
family with sequence similarity 50, member A
|
|
chr6_+_124125068
|
1.794
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr2_+_217498205
|
1.790
|
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr12_+_56075418
|
1.763
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr10_-_79397470
|
1.717
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr7_+_8008416
|
1.677
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr14_-_60097531
|
1.668
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr19_+_2476122
|
1.640
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr16_+_2039945
|
1.635
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr10_+_31610063
|
1.633
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr10_+_31608097
|
1.584
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr3_-_187463227
|
1.568
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr3_-_187463474
|
1.558
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr2_-_68547018
|
1.519
|
NM_001111101
NM_015463
|
CNRIP1
|
cannabinoid receptor interacting protein 1
|
|
chr8_-_108510094
|
1.510
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr19_+_36359346
|
1.510
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr6_+_72596405
|
1.498
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr16_-_73092533
|
1.482
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr2_+_109745996
|
1.429
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr5_+_119800004
|
1.407
|
NM_016644
|
PRR16
|
proline rich 16
|
|
chr6_+_72596648
|
1.386
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr16_+_1584161
|
1.377
|
NM_024600
|
TMEM204
|
transmembrane protein 204
|
|
chr5_-_14871555
|
1.331
|
NM_054027
|
ANKH
|
ankylosis, progressive homolog (mouse)
|
|
chr5_+_119799898
|
1.327
|
|
PRR16
|
proline rich 16
|
|
chr16_+_29823408
|
1.292
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chrX_-_154114431
|
1.288
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr13_-_44361032
|
1.281
|
NM_001242863
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr7_+_7222142
|
1.269
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr1_-_72748491
|
1.226
|
|
|
|
|
chr20_-_34025955
|
1.224
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr12_-_40499604
|
1.224
|
NM_052885
|
SLC2A13
|
solute carrier family 2 (facilitated glucose transporter), member 13
|
|
chr2_+_191745546
|
1.221
|
NM_014905
|
GLS
|
glutaminase
|
|
chr1_-_241520467
|
1.201
|
NM_002924
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr6_-_85473898
|
1.197
|
NM_001080508
|
TBX18
|
T-box 18
|
|
chr17_-_28619183
|
1.167
|
NM_000386
|
BLMH
|
bleomycin hydrolase
|
|
chrX_+_153672449
|
1.161
|
NM_004699
|
FAM50A
|
family with sequence similarity 50, member A
|
|
chr5_+_139060295
|
1.155
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr12_-_91576579
|
1.148
|
|
DCN
|
decorin
|
|
chr3_-_45267068
|
1.141
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr19_+_14551080
|
1.130
|
NM_213560
|
PKN1
|
protein kinase N1
|
|
chr2_+_23608297
|
1.128
|
NM_052920
|
KLHL29
|
kelch-like 29 (Drosophila)
|
|
chr11_+_113930430
|
1.127
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr1_+_183605207
|
1.111
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr12_-_91576805
|
1.102
|
NM_001920
|
DCN
|
decorin
|
|
chr14_-_60097314
|
1.094
|
|
RTN1
|
reticulon 1
|
|
chr14_+_100259445
|
1.092
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr10_-_79397290
|
1.089
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr10_+_60272868
|
1.056
|
NM_001080512
|
BICC1
|
bicaudal C homolog 1 (Drosophila)
|
|
chr5_+_40680021
|
1.045
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr20_-_34330093
|
1.040
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr14_-_60097223
|
1.037
|
|
RTN1
|
reticulon 1
|
|
chr7_+_100199881
|
1.036
|
NM_002593
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr4_-_57522469
|
1.014
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr8_-_92053043
|
1.009
|
NM_018710
|
TMEM55A
|
transmembrane protein 55A
|
|
chr4_-_89618928
|
1.004
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr16_+_69600110
|
1.001
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr15_+_63335086
|
1.000
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr1_-_241520384
|
0.987
|
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr8_-_60031545
|
0.971
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr18_-_25757351
|
0.967
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr11_-_70672644
|
0.954
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chrX_-_118827060
|
0.948
|
|
SEPT6
|
septin 6
|
|
chr10_-_79397546
|
0.929
|
NM_001014797
NM_001161352
NM_001161353
NM_002247
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr2_+_189839078
|
0.924
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr20_-_48099178
|
0.922
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr5_-_146833150
|
0.920
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr2_+_217498082
|
0.919
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr20_+_33146500
|
0.898
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr5_-_36151953
|
0.891
|
NM_001007527
|
LMBRD2
|
LMBR1 domain containing 2
|
|
chr5_+_137774897
|
0.876
|
|
REEP2
|
receptor accessory protein 2
|
|
chr2_+_191745791
|
0.873
|
|
GLS
|
glutaminase
|
|
chr5_+_176237435
|
0.873
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr4_+_15005364
|
0.870
|
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr1_+_163038564
|
0.869
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_-_24194760
|
0.865
|
NM_000147
|
FUCA1
|
fucosidase, alpha-L- 1, tissue
|
|
chrX_+_51486479
|
0.850
|
NM_018094
|
GSPT2
|
G1 to S phase transition 2
|
|
chr20_-_35374530
|
0.846
|
NM_022477
NM_032013
|
NDRG3
|
NDRG family member 3
|
|
chr17_-_8702438
|
0.845
|
NM_152599
|
MFSD6L
|
major facilitator superfamily domain containing 6-like
|
|
chr2_+_189839113
|
0.841
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr3_-_129325175
|
0.839
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr1_-_43232728
|
0.834
|
NM_001146289
NM_001243246
NM_022356
|
LEPRE1
|
leucine proline-enriched proteoglycan (leprecan) 1
|
|
chr6_-_139695349
|
0.832
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr13_+_32605436
|
0.818
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr17_-_28618836
|
0.812
|
|
BLMH
|
bleomycin hydrolase
|
|
chr17_+_61851512
|
0.811
|
|
|
|
|
chr5_-_146889618
|
0.811
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr22_-_21905372
|
0.811
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr1_+_64239566
|
0.807
|
NM_001083592
NM_005012
|
ROR1
|
receptor tyrosine kinase-like orphan receptor 1
|
|
chr16_+_29823557
|
0.806
|
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr14_-_23821423
|
0.802
|
NM_020372
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr2_-_202316227
|
0.801
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr3_+_49027770
|
0.800
|
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr19_-_47922468
|
0.797
|
|
MEIS3
|
Meis homeobox 3
|
|
chr7_-_27183225
|
0.797
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr10_-_32635842
|
0.783
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr1_-_43232656
|
0.780
|
|
LEPRE1
|
leucine proline-enriched proteoglycan (leprecan) 1
|
|
chr9_-_110251754
|
0.778
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr12_-_6665220
|
0.770
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr15_+_43809805
|
0.758
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr2_+_189839132
|
0.742
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr8_+_104383785
|
0.735
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr10_+_89419352
|
0.735
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr15_+_63334753
|
0.732
|
NM_000366
NM_001018004
NM_001018005
NM_001018006
NM_001018007
NM_001018020
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr1_+_51434231
|
0.731
|
NM_001262
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr19_-_18392436
|
0.716
|
|
JUND
|
jun D proto-oncogene
|
|
chr22_-_29075708
|
0.712
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr7_+_94024201
|
0.709
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr8_+_27183047
|
0.708
|
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr2_+_191745568
|
0.708
|
|
GLS
|
glutaminase
|
|
chr5_+_178450775
|
0.706
|
NM_001136116
|
ZNF879
|
zinc finger protein 879
|
|
chr19_-_40791224
|
0.705
|
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chrX_+_66766350
|
0.701
|
|
AR
|
androgen receptor
|
|
chr11_-_67188614
|
0.700
|
|
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme
|
|
chr2_+_24299751
|
0.699
|
|
LOC375190
|
UPF0638 protein B
|
|
chr6_-_111804413
|
0.699
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr5_+_137774632
|
0.698
|
NM_016606
|
REEP2
|
receptor accessory protein 2
|
|
chr5_-_137368682
|
0.696
|
|
FAM13B
|
family with sequence similarity 13, member B
|
|
chr1_+_178063302
|
0.696
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr8_+_104383701
|
0.693
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr20_+_34742656
|
0.692
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr16_+_2802634
|
0.692
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr3_+_32280211
|
0.689
|
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr5_-_159546389
|
0.684
|
NM_001130864
NM_052927
|
PWWP2A
|
PWWP domain containing 2A
|
|
chr4_+_174292100
|
0.684
|
|
SAP30
|
Sin3A-associated protein, 30kDa
|
|
chr5_+_139060058
|
0.683
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr1_-_182360400
|
0.680
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr19_-_31840118
|
0.679
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr1_-_57045129
|
0.675
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr13_+_49549953
|
0.675
|
NM_001079673
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr3_+_20081853
|
0.670
|
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr11_+_113931228
|
0.669
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr12_-_29936685
|
0.669
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr2_-_227663454
|
0.668
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr2_-_45236521
|
0.667
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr4_-_138453564
|
0.662
|
|
PCDH18
|
protocadherin 18
|
|
chr3_+_32280154
|
0.661
|
NM_178868
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr4_-_138453628
|
0.655
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr15_+_96873845
|
0.654
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr4_-_18023358
|
0.654
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr17_-_48264039
|
0.653
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr8_+_27183062
|
0.651
|
NM_173175
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr6_-_111804106
|
0.650
|
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr14_+_53019865
|
0.648
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr2_+_202316391
|
0.648
|
NM_001206864
NM_018571
|
STRADB
|
STE20-related kinase adaptor beta
|
|
chr12_-_63328663
|
0.647
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr21_-_34100210
|
0.646
|
|
SYNJ1
|
synaptojanin 1
|
|
chr4_+_174292027
|
0.646
|
NM_003864
|
SAP30
|
Sin3A-associated protein, 30kDa
|
|
chr20_+_35089817
|
0.642
|
NM_001042486
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr5_+_139060186
|
0.641
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr6_-_132272311
|
0.635
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr15_-_55880508
|
0.632
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr3_+_105085740
|
0.628
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr1_+_178063237
|
0.627
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr5_+_140602914
|
0.626
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr5_+_75699003
|
0.625
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr17_+_39240412
|
0.625
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr15_-_78369825
|
0.622
|
NM_015079
NM_144572
|
TBC1D2B
|
TBC1 domain family, member 2B
|
|
chr14_+_50087400
|
0.618
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr1_-_31845818
|
0.617
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr4_+_153457407
|
0.616
|
|
DKFZP434I0714
|
uncharacterized protein DKFZP434I0714
|
|
chr4_-_2263706
|
0.612
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr2_+_74741565
|
0.610
|
NM_016170
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr14_+_38678594
|
0.610
|
|
SSTR1
|
somatostatin receptor 1
|
|
chrX_-_154688260
|
0.609
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated 2
coagulation factor VIII-associated 3
coagulation factor VIII-associated 1
|
|
chr4_+_126237566
|
0.607
|
NM_024582
|
FAT4
|
FAT tumor suppressor homolog 4 (Drosophila)
|
|
chr8_+_27182995
|
0.604
|
NM_173176
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chrX_-_118827322
|
0.604
|
NM_015129
NM_145799
NM_145800
NM_145802
|
SEPT6
|
septin 6
|
|
chr5_-_137368778
|
0.603
|
NM_001101800
NM_001101801
NM_016603
|
FAM13B
|
family with sequence similarity 13, member B
|
|
chr1_-_92351613
|
0.602
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr8_-_57123827
|
0.598
|
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chrX_+_16964730
|
0.597
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr5_-_137089434
|
0.591
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr14_+_29236882
|
0.590
|
|
FOXG1
|
forkhead box G1
|
|
chr9_-_120177326
|
0.590
|
|
ASTN2
|
astrotactin 2
|
|
chr13_+_98086315
|
0.589
|
NM_021033
|
RAP2A
|
RAP2A, member of RAS oncogene family
|
|
chr2_-_198175494
|
0.589
|
NM_001195144
NM_153697
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr1_-_92351476
|
0.588
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr14_-_61115906
|
0.588
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr15_-_90294359
|
0.587
|
NM_018670
|
MESP1
|
mesoderm posterior 1 homolog (mouse)
|