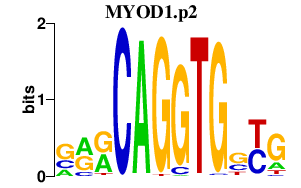
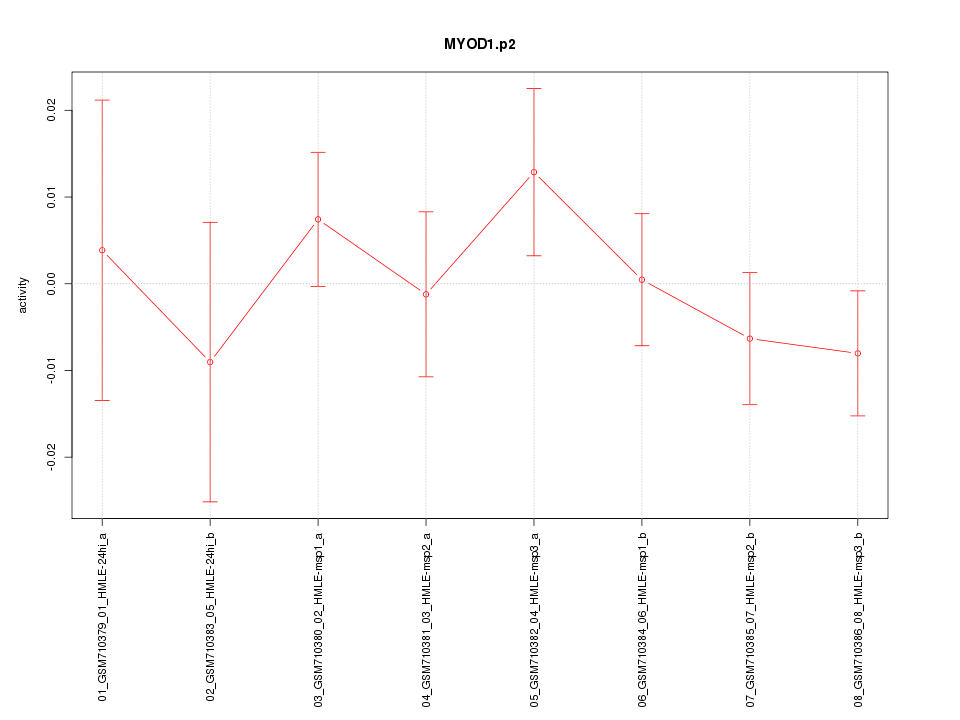
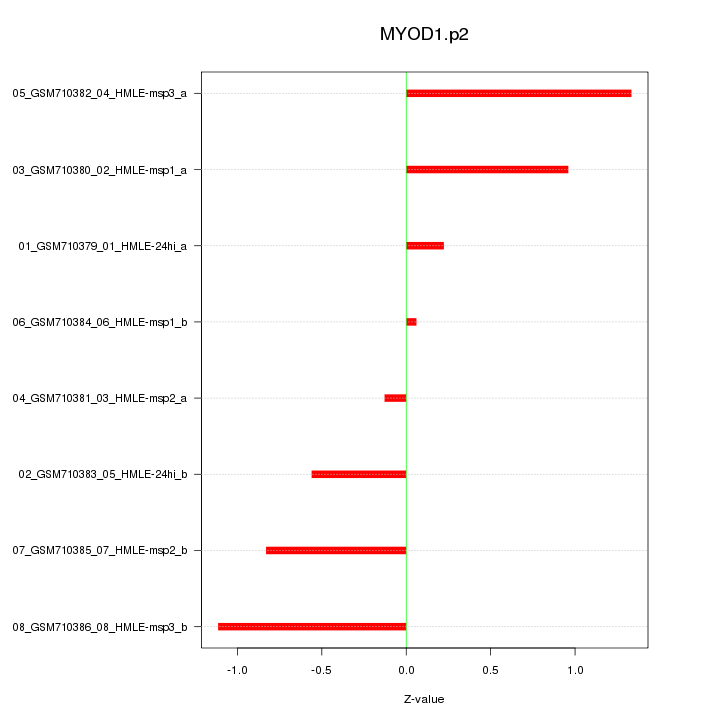
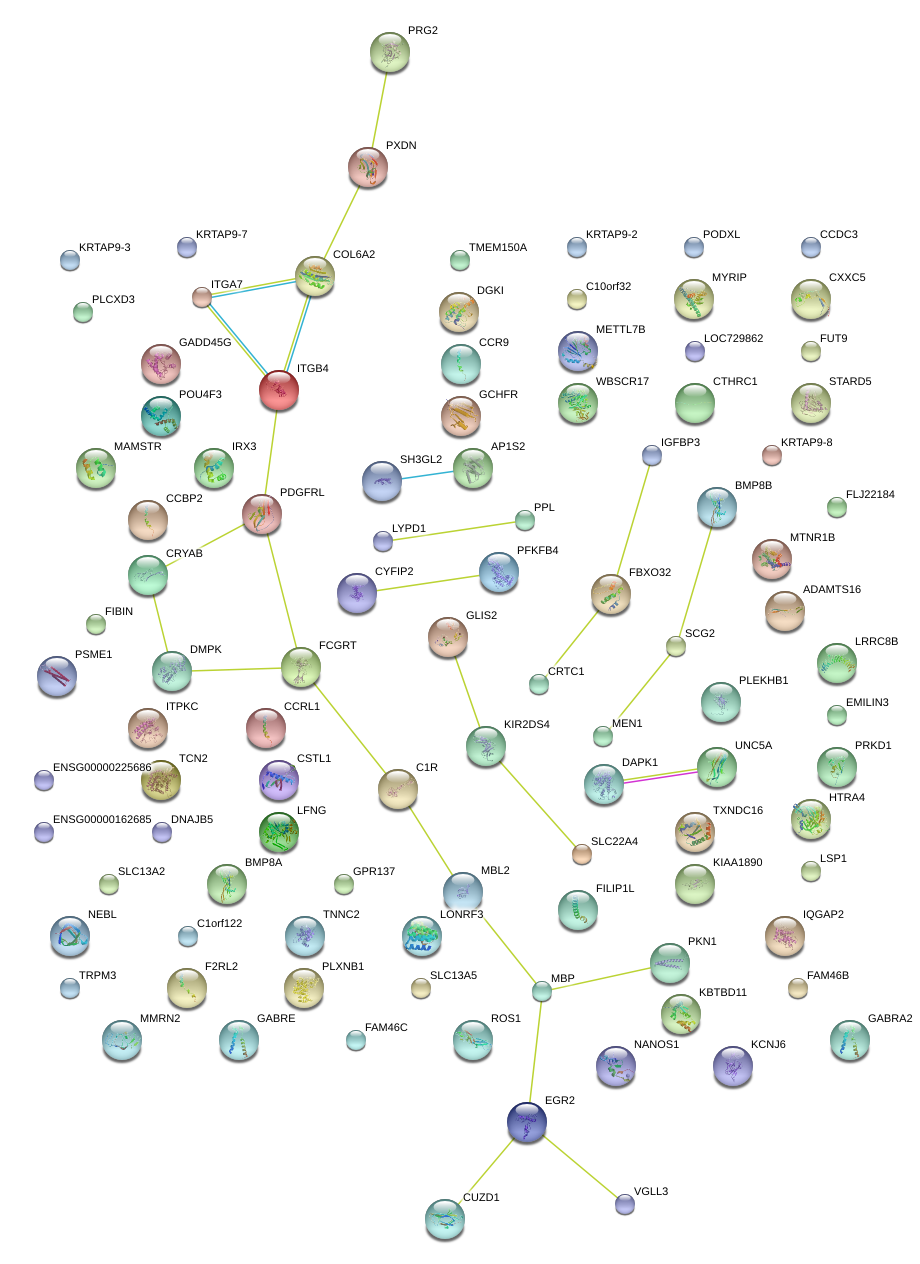
|
chr11_-_1769086
|
0.802
|
|
IFITM10
|
interferon induced transmembrane protein 10
|
|
chr5_+_95067511
|
0.383
|
|
|
|
|
chr17_+_39382878
|
0.366
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr9_+_17578952
|
0.306
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr21_+_47531369
|
0.300
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr10_+_120789227
|
0.292
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr19_-_49222956
|
0.277
|
NM_001130915
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr14_-_30396811
|
0.268
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr10_-_21462987
|
0.265
|
|
NEBL
|
nebulette
|
|
chr12_+_56075329
|
0.261
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr12_+_56075418
|
0.260
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr10_-_124605690
|
0.250
|
NM_022034
|
CUZD1
|
CUB and zona pellucida-like domains 1
|
|
chr19_-_7939325
|
0.247
|
NM_001190467
|
FLJ22184
|
putative uncharacterized protein FLJ22184
|
|
chr11_-_111782447
|
0.246
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr10_-_21463019
|
0.246
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr16_-_4987056
|
0.236
|
NM_002705
|
PPL
|
periplakin
|
|
chr1_+_89990396
|
0.234
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr7_-_45960738
|
0.231
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr17_+_73717556
|
0.224
|
|
ITGB4
|
integrin, beta 4
|
|
chr5_+_176237435
|
0.207
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chrX_+_118108595
|
0.203
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chrX_-_151143096
|
0.201
|
NM_004961
|
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon
|
|
chr19_+_41222915
|
0.195
|
NM_025194
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|
|
chrX_+_118108575
|
0.194
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr7_-_137531585
|
0.192
|
NM_004717
|
DGKI
|
diacylglycerol kinase, iota
|
|
chr15_+_41056243
|
0.188
|
NM_005258
|
GCHFR
|
GTP cyclohydrolase I feedback regulator
|
|
chr8_+_1922030
|
0.185
|
NM_014867
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr17_+_73717596
|
0.181
|
|
ITGB4
|
integrin, beta 4
|
|
chr19_+_50016435
|
0.181
|
NM_004107
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr11_+_92702788
|
0.181
|
NM_005959
|
MTNR1B
|
melatonin receptor 1B
|
|
chr2_-_224467049
|
0.179
|
|
SCG2
|
secretogranin II
|
|
chr19_+_41223282
|
0.179
|
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|
|
chr2_-_224467142
|
0.177
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr2_-_224467095
|
0.174
|
|
SCG2
|
secretogranin II
|
|
chr2_-_133427624
|
0.173
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr9_+_92219926
|
0.170
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr11_+_1874199
|
0.169
|
NM_002339
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr9_+_34990715
|
0.168
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr1_+_38273817
|
0.167
|
|
C1orf122
|
chromosome 1 open reading frame 122
|
|
chr10_+_104613966
|
0.166
|
NM_001136200
NM_144591
|
C10orf32-AS3MT
C10orf32
|
C10orf32-AS3MT readthrough
chromosome 10 open reading frame 32
|
|
chr10_-_64576074
|
0.165
|
|
EGR2
|
early growth response 2
|
|
chr12_-_7245006
|
0.164
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr17_+_39394269
|
0.163
|
NM_031963
|
KRTAP9-8
|
keratin associated protein 9-8
|
|
chr5_+_131630065
|
0.162
|
NM_003059
|
SLC22A4
|
solute carrier family 22 (organic cation/ergothioneine transporter), member 4
|
|
chr9_-_74061819
|
0.161
|
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr17_+_73717515
|
0.159
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr10_-_64576123
|
0.158
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr5_+_75699003
|
0.156
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr12_-_56101667
|
0.155
|
NM_001144996
NM_002206
|
ITGA7
|
integrin, alpha 7
|
|
chr18_-_74691951
|
0.155
|
|
MBP
|
myelin basic protein
|
|
chr5_+_145718586
|
0.154
|
NM_002700
|
POU4F3
|
POU class 4 homeobox 3
|
|
chr3_-_48470715
|
0.154
|
NM_002673
|
PLXNB1
|
plexin B1
|
|
chr12_-_56101465
|
0.152
|
|
ITGA7
|
integrin, alpha 7
|
|
chr6_-_117747017
|
0.149
|
NM_002944
|
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase
|
|
chr3_-_48594209
|
0.149
|
NM_004567
|
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
|
|
chr7_+_70597788
|
0.147
|
NM_022479
|
WBSCR17
|
Williams-Beuren syndrome chromosome region 17
|
|
chr11_+_73358593
|
0.146
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr5_+_139027868
|
0.144
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr3_-_99833348
|
0.144
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr9_+_90112754
|
0.143
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr6_+_96463844
|
0.143
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr19_+_18794487
|
0.142
|
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr22_+_31003069
|
0.141
|
NM_000355
NM_001184726
|
TCN2
|
transcobalamin II
|
|
chr19_+_14544216
|
0.141
|
|
PKN1
|
protein kinase N1
|
|
chr8_+_104383701
|
0.141
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr8_+_72756218
|
0.140
|
|
LOC100132891
|
uncharacterized LOC100132891
|
|
chr3_-_87040246
|
0.140
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr16_-_54318889
|
0.139
|
|
IRX3
|
iroquois homeobox 3
|
|
chr8_+_72756335
|
0.138
|
|
|
|
|
chr7_+_2557496
|
0.136
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr20_+_23420321
|
0.135
|
NM_138283
|
CSTL1
|
cystatin-like 1
|
|
chr1_-_40254510
|
0.135
|
|
BMP8B
|
bone morphogenetic protein 8b
|
|
chr5_+_5140442
|
0.133
|
NM_139056
|
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16
|
|
chr10_-_13043570
|
0.133
|
NM_031455
|
CCDC3
|
coiled-coil domain containing 3
|
|
chr1_+_118148506
|
0.133
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr5_+_156693051
|
0.132
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr3_+_45927995
|
0.131
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr2_-_133428480
|
0.131
|
NM_144586
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr2_-_85829757
|
0.129
|
NM_001031738
|
TMEM150A
|
transmembrane protein 150A
|
|
chr11_+_27015833
|
0.128
|
|
FIBIN
|
fin bud initiation factor homolog (zebrafish)
|
|
chr1_-_27339326
|
0.128
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr8_+_38831667
|
0.127
|
NM_153692
|
HTRA4
|
HtrA serine peptidase 4
|
|
chr3_+_39851028
|
0.126
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr16_+_4385141
|
0.126
|
|
GLIS2
|
GLIS family zinc finger 2
|
|
chrX_-_15872938
|
0.124
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr8_+_17433941
|
0.124
|
NM_006207
|
PDGFRL
|
platelet-derived growth factor receptor-like
|
|
chrX_-_15872871
|
0.124
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr21_-_39288661
|
0.122
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr3_-_48471401
|
0.121
|
NM_001130082
|
PLXNB1
|
plexin B1
|
|
chr20_-_39995497
|
0.120
|
NM_052846
|
EMILIN3
|
elastin microfibril interfacer 3
|
|
chr5_-_41510627
|
0.120
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr14_+_24605366
|
0.119
|
NM_006263
NM_176783
|
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha)
|
|
chr19_-_46285746
|
0.119
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr17_+_26800657
|
0.117
|
NM_001145975
NM_001145976
NM_003984
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2
|
|
chr4_-_46391393
|
0.117
|
NM_001114175
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2
|
|
chr5_-_75919051
|
0.117
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr7_-_131241321
|
0.116
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr1_-_27339448
|
0.116
|
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr8_-_124553253
|
0.116
|
|
FBXO32
|
F-box protein 32
|
|
chr15_-_81616439
|
0.115
|
NM_181900
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5
|
|
chr20_-_44455932
|
0.115
|
NM_003279
|
TNNC2
|
troponin C type 2 (fast)
|
|
chr6_+_168841829
|
0.115
|
NM_001166412
NM_022138
|
SMOC2
|
SPARC related modular calcium binding 2
|
|
chr2_-_190044330
|
0.114
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr4_-_142054431
|
0.114
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr17_+_39405938
|
0.113
|
NM_033191
|
KRTAP9-4
|
keratin associated protein 9-4
|
|
chr19_+_50016606
|
0.113
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr7_-_124405029
|
0.113
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chrX_+_118109314
|
0.113
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr1_+_27668499
|
0.113
|
|
SYTL1
|
synaptotagmin-like 1
|
|
chr6_-_24911194
|
0.113
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr13_-_26625164
|
0.112
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr5_-_66492574
|
0.112
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr11_-_77899640
|
0.111
|
NM_001029859
|
KCTD21
|
potassium channel tetramerisation domain containing 21
|
|
chr10_-_98480174
|
0.111
|
NM_152309
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr2_+_217498082
|
0.110
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr5_+_139028483
|
0.110
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr22_-_21905372
|
0.110
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr4_-_159092509
|
0.110
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr1_-_145075636
|
0.109
|
NM_022359
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr2_+_217498205
|
0.108
|
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr1_-_168698412
|
0.107
|
NM_001937
|
DPT
|
dermatopontin
|
|
chr19_-_46285645
|
0.107
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr4_-_46391771
|
0.107
|
NM_000807
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2
|
|
chr19_-_39368889
|
0.106
|
NM_001195833
NM_198445
|
RINL
|
Ras and Rab interactor-like
|
|
chr20_+_44519590
|
0.106
|
NM_000308
NM_001127695
NM_001167594
|
CTSA
|
cathepsin A
|
|
chr20_+_54933982
|
0.106
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr4_+_134073180
|
0.105
|
|
PCDH10
|
protocadherin 10
|
|
chr12_+_90102639
|
0.104
|
|
LOC338758
|
uncharacterized LOC338758
|
|
chr2_+_205410465
|
0.104
|
NM_057177
NM_152526
NM_205863
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr3_-_49851356
|
0.104
|
NM_003335
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr5_+_137774632
|
0.104
|
NM_016606
|
REEP2
|
receptor accessory protein 2
|
|
chr5_+_137774897
|
0.103
|
|
REEP2
|
receptor accessory protein 2
|
|
chr19_-_6375859
|
0.103
|
NM_004158
|
PSPN
|
persephin
|
|
chr17_-_7017971
|
0.103
|
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr1_+_53925482
|
0.102
|
|
DMRTB1
|
DMRT-like family B with proline-rich C-terminal, 1
|
|
chr14_+_24605393
|
0.102
|
|
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha)
|
|
chr6_+_168841873
|
0.102
|
|
SMOC2
|
SPARC related modular calcium binding 2
|
|
chr15_+_81489218
|
0.102
|
NM_001172128
|
IL16
|
interleukin 16
|
|
chr19_+_36359346
|
0.102
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr18_-_56940616
|
0.101
|
NM_013435
|
RAX
|
retina and anterior neural fold homeobox
|
|
chr3_-_49851217
|
0.101
|
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr7_+_44788453
|
0.101
|
NM_031449
|
ZMIZ2
|
zinc finger, MIZ-type containing 2
|
|
chr11_+_1886394
|
0.100
|
NM_001242932
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr7_-_150864866
|
0.099
|
NM_001098834
|
GBX1
|
gastrulation brain homeobox 1
|
|
chr22_-_31885512
|
0.099
|
NM_001164501
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1
|
|
chr8_-_124553448
|
0.099
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr3_-_9920633
|
0.098
|
NM_022094
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr6_-_6007632
|
0.098
|
NM_016588
|
NRN1
|
neuritin 1
|
|
chr17_+_5972425
|
0.098
|
|
WSCD1
|
WSC domain containing 1
|
|
chr3_-_9920471
|
0.098
|
NM_001199623
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr6_+_31683084
|
0.097
|
NM_021246
|
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D
|
|
chr3_+_105085740
|
0.097
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr5_-_159739482
|
0.097
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr3_+_8543539
|
0.097
|
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr5_+_139027955
|
0.096
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr5_+_139027907
|
0.096
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr10_-_15413054
|
0.096
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr8_+_104383785
|
0.096
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr5_+_96271086
|
0.096
|
|
LNPEP
|
leucyl/cystinyl aminopeptidase
|
|
chr12_-_52585686
|
0.096
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chrX_-_13956530
|
0.095
|
|
GPM6B
|
glycoprotein M6B
|
|
chr6_+_35182182
|
0.095
|
NM_152753
|
SCUBE3
|
signal peptide, CUB domain, EGF-like 3
|
|
chr6_+_35181721
|
0.095
|
|
SCUBE3
|
signal peptide, CUB domain, EGF-like 3
|
|
chr19_+_18111923
|
0.094
|
NM_001025604
|
ARRDC2
|
arrestin domain containing 2
|
|
chr19_-_11591439
|
0.094
|
NM_001420
NM_032281
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr3_+_170136652
|
0.094
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr5_+_149569810
|
0.094
|
|
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7
|
|
chr19_+_8478181
|
0.094
|
NM_001005415
NM_001005416
NM_016496
|
MARCH2
|
membrane-associated ring finger (C3HC4) 2
|
|
chr12_-_7245048
|
0.094
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr8_-_57123827
|
0.094
|
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr1_+_210501595
|
0.094
|
NM_001122834
NM_001170564
|
HHAT
|
hedgehog acyltransferase
|
|
chr4_-_2366530
|
0.093
|
NM_001172659
NM_001172660
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chr2_+_109745996
|
0.093
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr19_-_46285752
|
0.092
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chrX_-_19140581
|
0.092
|
NM_001079858
NM_001079859
NM_001079860
NM_001184833
NM_001184834
NM_001184835
NM_001184836
NM_001184837
NM_005756
|
GPR64
|
G protein-coupled receptor 64
|
|
chr1_-_183387501
|
0.092
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr13_+_102104941
|
0.092
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr1_+_77747602
|
0.091
|
NM_174858
|
AK5
|
adenylate kinase 5
|
|
chr17_+_77030471
|
0.091
|
NM_198593
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr17_-_72542148
|
0.091
|
|
CD300C
|
CD300c molecule
|
|
chrX_-_152939256
|
0.091
|
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chrX_-_100306859
|
0.091
|
|
TRMT2B
|
TRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae)
|
|
chr21_-_45079327
|
0.090
|
NM_007031
|
HSF2BP
|
heat shock transcription factor 2 binding protein
|
|
chr3_+_69788562
|
0.090
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_+_53925071
|
0.090
|
NM_033067
|
DMRTB1
|
DMRT-like family B with proline-rich C-terminal, 1
|
|
chr4_-_16085593
|
0.090
|
NM_001145847
NM_001145848
|
PROM1
|
prominin 1
|
|
chr10_-_13276327
|
0.089
|
NM_145314
|
UCMA
|
upper zone of growth plate and cartilage matrix associated
|
|
chr4_-_129208964
|
0.089
|
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr1_-_26232890
|
0.089
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr1_+_23695441
|
0.089
|
|
C1orf213
|
chromosome 1 open reading frame 213
|
|
chr13_+_32605436
|
0.088
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr4_-_57976550
|
0.088
|
NM_001253835
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chrX_-_15872882
|
0.088
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr11_-_1785087
|
0.088
|
|
CTSD
|
cathepsin D
|
|
chr5_-_126366439
|
0.088
|
NM_178450
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3
|
|
chr17_+_78194090
|
0.088
|
NM_001166347
NM_001166348
NM_001166349
NM_173626
|
SLC26A11
|
solute carrier family 26, member 11
|
|
chr19_+_50016490
|
0.088
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr14_-_61115906
|
0.087
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr9_-_115652961
|
0.087
|
NM_033051
|
SLC46A2
|
solute carrier family 46, member 2
|
|
chr6_+_26501448
|
0.087
|
NM_001732
|
BTN1A1
|
butyrophilin, subfamily 1, member A1
|