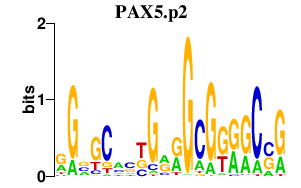
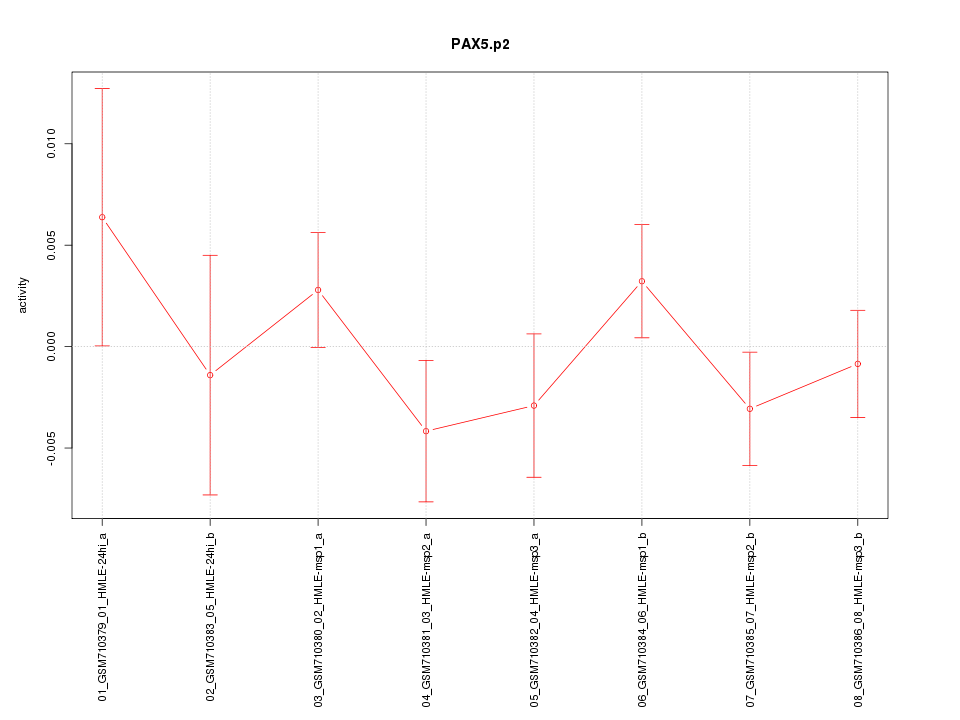
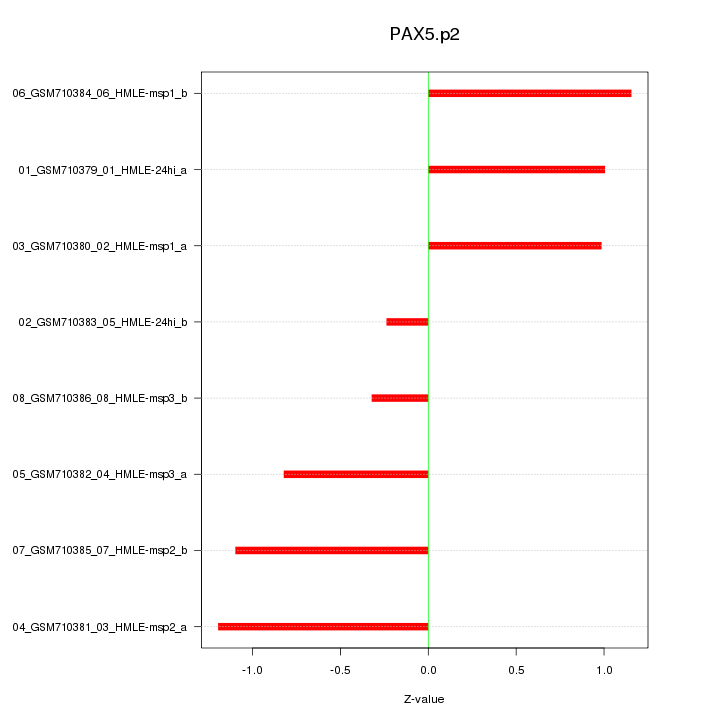
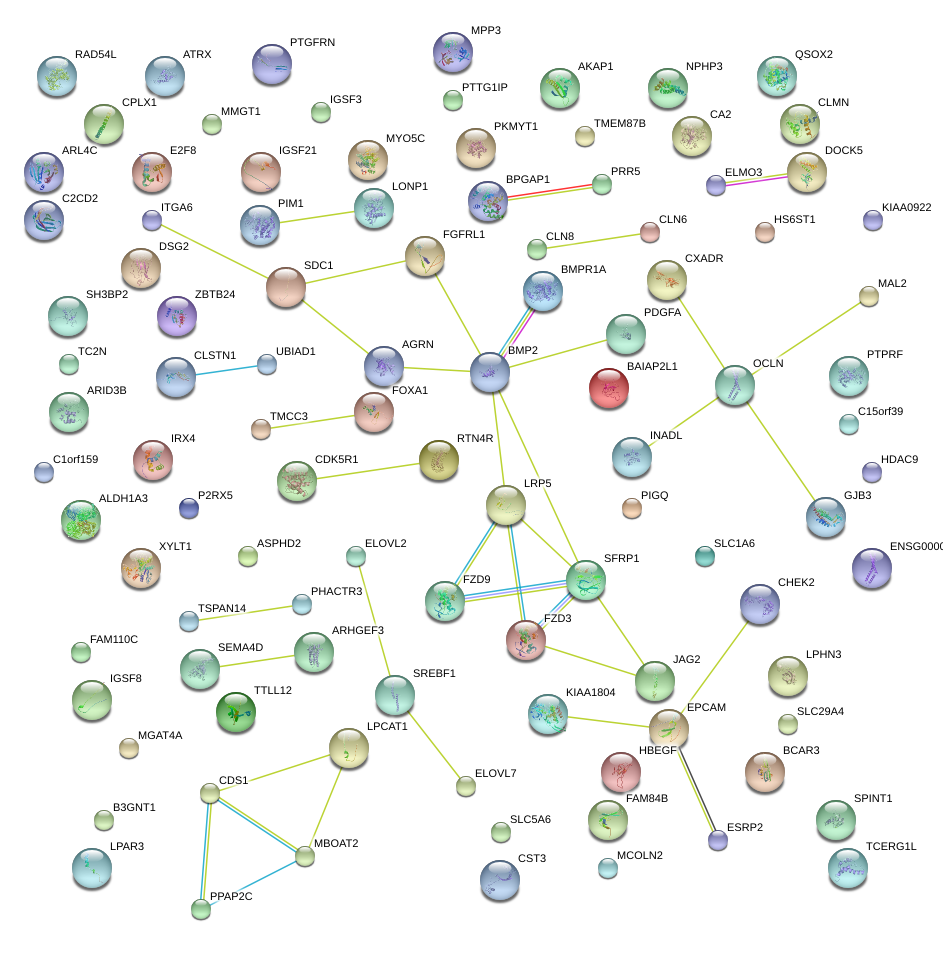
|
chr15_+_101420004
|
0.767
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr2_-_46384
|
0.753
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr19_-_291168
|
0.731
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr1_-_1051458
|
0.617
|
|
C1orf159
|
chromosome 1 open reading frame 159
|
|
chr19_-_291335
|
0.554
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr15_+_101420032
|
0.549
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr3_-_132441216
|
0.529
|
|
NPHP3
|
nephronophthisis 3 (adolescent)
|
|
chr6_-_109804322
|
0.512
|
|
ZBTB24
|
zinc finger and BTB domain containing 24
|
|
chr14_-_105634708
|
0.503
|
|
JAG2
|
jagged 2
|
|
chr22_-_20255614
|
0.441
|
NM_023004
|
RTN4R
|
reticulon 4 receptor
|
|
chr16_-_68269948
|
0.440
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr22_-_43583132
|
0.437
|
NM_015140
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12
|
|
chr1_+_117452359
|
0.433
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr5_-_60140041
|
0.431
|
NM_001104558
NM_024930
|
ELOVL7
|
ELOVL fatty acid elongase 7
|
|
chr2_+_47596286
|
0.426
|
NM_002354
|
EPCAM
|
epithelial cell adhesion molecule
|
|
chr2_+_47596447
|
0.413
|
|
EPCAM
|
epithelial cell adhesion molecule
|
|
chr16_-_17564737
|
0.405
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr17_+_30813928
|
0.403
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr6_-_109804432
|
0.391
|
NM_001164313
NM_014797
|
ZBTB24
|
zinc finger and BTB domain containing 24
|
|
chr4_-_819879
|
0.388
|
NM_006651
|
CPLX1
|
complexin 1
|
|
chr1_+_11333244
|
0.380
|
NM_013319
|
UBIAD1
|
UbiA prenyltransferase domain containing 1
|
|
chr17_+_55162530
|
0.380
|
NM_003488
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr2_+_112812799
|
0.372
|
NM_032824
|
TMEM87B
|
transmembrane protein 87B
|
|
chr3_-_57113292
|
0.361
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr17_-_41910439
|
0.352
|
NM_001932
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3)
|
|
chr9_-_139137646
|
0.351
|
NM_181701
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2
|
|
chr22_+_45148437
|
0.350
|
NM_001017526
NM_001198726
NM_181335
|
ARHGAP8
|
Rho GTPase activating protein 8
|
|
chr12_-_95044205
|
0.348
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr14_-_92302612
|
0.347
|
NM_001128595
NM_152332
|
TC2N
|
tandem C2 domains, nuclear
|
|
chr10_+_82213892
|
0.346
|
NM_001128309
NM_030927
|
TSPAN14
|
tetraspanin 14
|
|
chr15_-_52587851
|
0.338
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr14_-_95786095
|
0.335
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr6_+_37137882
|
0.334
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chrX_-_135056132
|
0.331
|
NM_173470
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr2_-_27435014
|
0.330
|
NM_021095
|
SLC5A6
|
solute carrier family 5 (sodium-dependent vitamin transporter), member 6
|
|
chr1_-_160068359
|
0.329
|
NM_052868
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr15_-_68521921
|
0.329
|
NM_017882
|
CLN6
|
ceroid-lipofuscinosis, neuronal 6, late infantile, variant
|
|
chr8_+_86376130
|
0.325
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr15_+_74833517
|
0.320
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr20_+_6748744
|
0.320
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr1_-_85358851
|
0.315
|
NM_012152
|
LPAR3
|
lysophosphatidic acid receptor 3
|
|
chr20_+_58152563
|
0.311
|
NM_001199505
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr22_+_45098091
|
0.306
|
NM_181334
|
PRR5-ARHGAP8
PRR5
|
PRR5-ARHGAP8 readthrough
proline rich 5 (renal)
|
|
chr15_+_41136178
|
0.305
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr1_-_117210301
|
0.302
|
NM_001007237
NM_001542
|
IGSF3
|
immunoglobulin superfamily, member 3
|
|
chr1_-_160068617
|
0.301
|
NM_001206665
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr1_+_955468
|
0.301
|
NM_198576
|
AGRN
|
agrin
|
|
chr5_+_68788387
|
0.300
|
NM_001205254
|
OCLN
|
occludin
|
|
chr1_-_175161832
|
0.297
|
NM_001162895
NM_014656
NM_001162893
NM_001162894
|
KIAA0040
|
KIAA0040
|
|
chr9_-_92112887
|
0.297
|
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D
|
|
chr10_+_88516340
|
0.297
|
NM_004329
|
BMPR1A
|
bone morphogenetic protein receptor, type IA
|
|
chr4_+_154387450
|
0.296
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr22_-_43583058
|
0.295
|
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12
|
|
chr22_+_26825220
|
0.294
|
NM_020437
|
ASPHD2
|
aspartate beta-hydroxylase domain containing 2
|
|
chr4_+_2794811
|
0.291
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr21_+_18885296
|
0.289
|
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr10_+_82214066
|
0.284
|
|
TSPAN14
|
tetraspanin 14
|
|
chr1_+_62208091
|
0.281
|
NM_176877
|
INADL
|
InaD-like (Drosophila)
|
|
chr2_-_235405692
|
0.279
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr1_-_160068261
|
0.279
|
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr1_+_46713433
|
0.278
|
|
RAD54L
|
RAD54-like (S. cerevisiae)
|
|
chr8_-_41166931
|
0.275
|
NM_003012
|
SFRP1
|
secreted frizzled-related protein 1
|
|
chr1_+_46713385
|
0.273
|
|
RAD54L
|
RAD54-like (S. cerevisiae)
|
|
chr7_-_98030165
|
0.272
|
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
chr1_+_233463513
|
0.272
|
NM_032435
|
KIAA1804
|
mixed lineage kinase 4
|
|
chr1_+_43996478
|
0.270
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr8_-_127570710
|
0.270
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr4_+_85504056
|
0.269
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr1_+_46713359
|
0.266
|
NM_001142548
NM_003579
|
RAD54L
|
RAD54-like (S. cerevisiae)
|
|
chr17_-_3599298
|
0.265
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr5_-_1524022
|
0.265
|
NM_024830
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1
|
|
chr10_+_82214071
|
0.264
|
|
TSPAN14
|
tetraspanin 14
|
|
chr1_+_35247838
|
0.262
|
NM_001005752
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr1_-_48462561
|
0.260
|
NM_001194986
|
LOC388630
|
UPF0632 protein A
|
|
chr8_+_25042066
|
0.258
|
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr17_-_17726855
|
0.257
|
|
SREBF1
|
sterol regulatory element binding transcription factor 1
|
|
chr17_-_3599359
|
0.256
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr11_-_19263144
|
0.256
|
|
E2F8
|
E2F transcription factor 8
|
|
chr14_-_38064176
|
0.256
|
|
FOXA1
|
forkhead box A1
|
|
chr1_-_85462622
|
0.254
|
NM_153259
|
MCOLN2
|
mucolipin 2
|
|
chr2_-_20424627
|
0.249
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr5_-_1882764
|
0.249
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr11_-_19262424
|
0.249
|
NM_024680
|
E2F8
|
E2F transcription factor 8
|
|
chr14_-_105635114
|
0.248
|
NM_002226
NM_145159
|
JAG2
|
jagged 2
|
|
chr8_+_1712011
|
0.247
|
|
CLN8
|
ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation)
|
|
chr19_-_15090349
|
0.247
|
|
SLC1A6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6
|
|
chr4_+_1005395
|
0.244
|
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr16_+_67233013
|
0.242
|
NM_024712
|
ELMO3
|
engulfment and cell motility 3
|
|
chr16_-_3030391
|
0.242
|
|
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1
|
|
chr21_+_47649066
|
0.242
|
|
MCM3AP-AS1
|
MCM3AP antisense RNA 1 (non-protein coding)
|
|
chr5_-_139726173
|
0.237
|
NM_001945
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr15_+_75494235
|
0.237
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr11_+_68079999
|
0.237
|
NM_002335
|
LRP5
|
low density lipoprotein receptor-related protein 5
|
|
chr10_-_133109983
|
0.236
|
NM_174937
|
TCERG1L
|
transcription elongation regulator 1-like
|
|
chr2_+_173292306
|
0.235
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr8_+_120220554
|
0.235
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr2_-_9143742
|
0.234
|
NM_138799
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr21_-_43373938
|
0.232
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr2_-_20425166
|
0.229
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr1_+_955609
|
0.229
|
|
AGRN
|
agrin
|
|
chr2_-_99347430
|
0.228
|
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr21_+_18885104
|
0.228
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr2_+_62423260
|
0.226
|
NM_006577
|
B3GNT2
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2
|
|
chr7_-_559479
|
0.226
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr1_+_11333502
|
0.225
|
|
UBIAD1
|
UbiA prenyltransferase domain containing 1
|
|
chr1_-_94147201
|
0.222
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr2_+_173292344
|
0.221
|
|
ITGA6
|
integrin, alpha 6
|
|
chr21_-_46293598
|
0.221
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr7_+_18126543
|
0.221
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr20_-_23618382
|
0.220
|
NM_000099
|
CST3
|
cystatin C
|
|
chr14_-_38064440
|
0.217
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr21_-_46293609
|
0.217
|
NM_004339
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr21_-_46293532
|
0.214
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr10_+_82214074
|
0.212
|
|
TSPAN14
|
tetraspanin 14
|
|
chr7_+_5322553
|
0.212
|
NM_001040661
NM_153247
|
SLC29A4
|
solute carrier family 29 (nucleoside transporters), member 4
|
|
chr1_+_18434236
|
0.211
|
NM_032880
|
IGSF21
|
immunoglobin superfamily, member 21
|
|
chr6_-_11044354
|
0.211
|
NM_017770
|
ELOVL2
|
ELOVL fatty acid elongase 2
|
|
chr2_-_129076045
|
0.209
|
NM_004807
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1
|
|
chr18_+_29078026
|
0.209
|
NM_001943
|
DSG2
|
desmoglein 2
|
|
chr4_+_1005609
|
0.209
|
NM_001004356
NM_001004358
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr1_-_9884024
|
0.208
|
|
CLSTN1
|
calsyntenin 1
|
|
chr8_+_28351694
|
0.208
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr7_-_105752718
|
0.208
|
NM_182715
|
SYPL1
|
synaptophysin-like 1
|
|
chr20_-_56284945
|
0.208
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr10_-_100027943
|
0.206
|
NM_032211
|
LOXL4
|
lysyl oxidase-like 4
|
|
chr6_-_99395774
|
0.205
|
|
FBXL4
|
F-box and leucine-rich repeat protein 4
|
|
chr21_-_46293561
|
0.205
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr22_-_50946020
|
0.203
|
|
LMF2
|
lipase maturation factor 2
|
|
chr5_-_77944341
|
0.203
|
NM_005779
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr2_+_148602050
|
0.203
|
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr1_+_210111575
|
0.203
|
|
SYT14
|
synaptotagmin XIV
|
|
chr6_+_138188352
|
0.203
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr19_+_45312294
|
0.202
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr20_+_61273786
|
0.201
|
NM_016354
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr20_+_61273823
|
0.201
|
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr15_+_75494233
|
0.199
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr8_+_25042254
|
0.198
|
NM_024940
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr2_-_198175324
|
0.198
|
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr21_+_46293925
|
0.197
|
|
|
|
|
chr6_-_138428477
|
0.196
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr3_+_113666544
|
0.195
|
NM_173570
|
ZDHHC23
|
zinc finger, DHHC-type containing 23
|
|
chr1_-_202936333
|
0.195
|
NM_016243
|
CYB5R1
|
cytochrome b5 reductase 1
|
|
chr6_+_14117857
|
0.194
|
NM_001040280
NM_004233
|
CD83
|
CD83 molecule
|
|
chr6_+_4776826
|
0.193
|
|
CDYL
|
chromodomain protein, Y-like
|
|
chr6_+_4136242
|
0.192
|
|
LOC100507506
|
uncharacterized LOC100507506
|
|
chr1_+_227058241
|
0.191
|
NM_000447
NM_012486
|
PSEN2
|
presenilin 2 (Alzheimer disease 4)
|
|
chr7_-_138665853
|
0.190
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr3_-_46735133
|
0.190
|
NM_001190707
NM_147129
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr13_+_26042965
|
0.189
|
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2
|
|
chr15_+_90744506
|
0.189
|
NM_198925
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr22_-_50946129
|
0.189
|
NM_033200
|
LMF2
|
lipase maturation factor 2
|
|
chr1_+_222791151
|
0.189
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr1_-_9884549
|
0.188
|
NM_001009566
NM_014944
|
CLSTN1
|
calsyntenin 1
|
|
chr3_-_124931562
|
0.188
|
NM_024628
|
SLC12A8
|
solute carrier family 12 (potassium/chloride transporters), member 8
|
|
chr8_-_144650998
|
0.188
|
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr1_+_44584521
|
0.187
|
NM_173484
|
KLF17
|
Kruppel-like factor 17
|
|
chr17_-_74497406
|
0.187
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chrX_-_152989822
|
0.187
|
NM_005745
|
BCAP31
|
B-cell receptor-associated protein 31
|
|
chr17_-_3599487
|
0.186
|
NM_001204519
NM_001204520
NM_002561
NM_175080
|
P2RX5-TAX1BP3
P2RX5
|
P2RX5-TAX1BP3 readthrough
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr15_+_73976621
|
0.184
|
NM_001024736
NM_025240
|
CD276
|
CD276 molecule
|
|
chr22_+_45064323
|
0.184
|
NM_001017528
NM_001017529
|
PRR5
|
proline rich 5 (renal)
|
|
chr16_+_67465019
|
0.183
|
NM_000196
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2
|
|
chr6_-_99395807
|
0.183
|
NM_012160
|
FBXL4
|
F-box and leucine-rich repeat protein 4
|
|
chr21_-_16437125
|
0.183
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr10_-_116164514
|
0.182
|
NM_001001936
NM_032550
|
AFAP1L2
|
actin filament associated protein 1-like 2
|
|
chr3_+_31574431
|
0.181
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr15_+_75494196
|
0.181
|
NM_015492
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr4_-_37687932
|
0.181
|
NM_001085399
NM_001085400
|
RELL1
|
RELT-like 1
|
|
chr1_-_11714738
|
0.179
|
NM_012168
|
FBXO2
|
F-box protein 2
|
|
chr11_-_113345841
|
0.179
|
NM_000795
NM_016574
|
DRD2
|
dopamine receptor D2
|
|
chr6_+_12012723
|
0.178
|
NM_002114
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1
|
|
chr19_-_7939325
|
0.176
|
NM_001190467
|
FLJ22184
|
putative uncharacterized protein FLJ22184
|
|
chr17_-_31203889
|
0.176
|
NM_015194
|
MYO1D
|
myosin ID
|
|
chr4_+_106816593
|
0.176
|
NM_001033047
NM_001184690
NM_001184691
NM_001184692
NM_001184693
|
NPNT
|
nephronectin
|
|
chr13_+_26042917
|
0.175
|
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2
|
|
chr2_+_233562042
|
0.175
|
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr1_-_53793743
|
0.174
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr19_+_30302861
|
0.174
|
NM_001238
|
CCNE1
|
cyclin E1
|
|
chr1_+_116519175
|
0.174
|
|
SLC22A15
|
solute carrier family 22, member 15
|
|
chr6_-_39197225
|
0.173
|
NM_003740
|
KCNK5
|
potassium channel, subfamily K, member 5
|
|
chr4_+_103422730
|
0.173
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr6_+_14118002
|
0.172
|
|
CD83
|
CD83 molecule
|
|
chr1_+_65613211
|
0.171
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr20_+_62328068
|
0.170
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr16_-_87903028
|
0.170
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr9_+_22447064
|
0.170
|
|
|
|
|
chr15_+_41136622
|
0.169
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr7_-_105752650
|
0.167
|
|
SYPL1
|
synaptophysin-like 1
|
|
chr2_-_70781146
|
0.166
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr22_+_29279573
|
0.165
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr9_-_100954883
|
0.165
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr9_-_23821808
|
0.163
|
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr7_-_143105897
|
0.162
|
NM_005232
|
EPHA1
|
EPH receptor A1
|
|
chr20_+_59827452
|
0.162
|
NM_001794
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal)
|
|
chr3_-_133748828
|
0.162
|
NM_005630
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chr1_+_955536
|
0.162
|
|
|
|
|
chr3_-_123603144
|
0.162
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr11_+_121322848
|
0.161
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr18_+_47088400
|
0.160
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr7_+_26331611
|
0.160
|
|
SNX10
|
sorting nexin 10
|