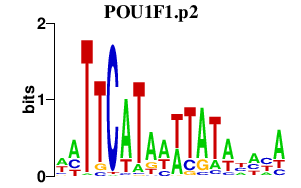
|
chr12_-_91573264
|
0.917
|
NM_133503
|
DCN
|
decorin
|
|
chr5_-_75919051
|
0.870
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr6_+_72922513
|
0.794
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr3_+_12392993
|
0.691
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chrX_+_66788682
|
0.620
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr6_+_72926462
|
0.605
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_-_57045214
|
0.588
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr3_+_148447966
|
0.587
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr11_-_102401430
|
0.528
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chr2_+_210444364
|
0.455
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr5_+_156696351
|
0.343
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr12_-_10573041
|
0.336
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chr1_+_114522029
|
0.335
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr12_+_55248298
|
0.331
|
NM_058173
|
MUCL1
|
mucin-like 1
|
|
chr12_+_21679190
|
0.325
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr12_-_91576579
|
0.314
|
|
DCN
|
decorin
|
|
chr8_-_19460005
|
0.313
|
NM_001130518
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1
|
|
chr2_+_109204777
|
0.303
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr20_-_45061554
|
0.301
|
|
ELMO2
|
engulfment and cell motility 2
|
|
chr4_+_70894129
|
0.299
|
NM_000200
|
HTN3
|
histatin 3
|
|
chr13_-_38172862
|
0.293
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr2_+_102770401
|
0.289
|
NM_000877
|
IL1R1
|
interleukin 1 receptor, type I
|
|
chr1_-_57045235
|
0.286
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr3_-_121379757
|
0.284
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr4_-_100242487
|
0.276
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr4_+_74347461
|
0.254
|
NM_001133
|
AFM
|
afamin
|
|
chr3_-_121379701
|
0.253
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr10_-_21186530
|
0.250
|
NM_006393
|
NEBL
|
nebulette
|
|
chr1_-_163172863
|
0.237
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr5_+_92918924
|
0.227
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr8_-_82395401
|
0.223
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr2_+_24807345
|
0.216
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr14_-_73453651
|
0.214
|
NM_178441
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr14_-_101036130
|
0.213
|
NM_020836
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr18_+_6834431
|
0.210
|
NM_001010000
|
ARHGAP28
|
Rho GTPase activating protein 28
|
|
chr19_-_44384240
|
0.209
|
NM_001033719
|
ZNF404
|
zinc finger protein 404
|
|
chr18_-_3011944
|
0.208
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr15_-_99789585
|
0.208
|
NM_001040655
NM_001040656
NM_001040657
NM_001040658
NM_001040659
NM_001040660
NM_022905
|
TTC23
|
tetratricopeptide repeat domain 23
|
|
chr2_-_88427535
|
0.206
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr9_-_16727887
|
0.204
|
|
BNC2
|
basonuclin 2
|
|
chr4_+_71108304
|
0.202
|
NM_005212
|
CSN3
|
casein kappa
|
|
chr9_-_123812535
|
0.202
|
NM_001735
|
C5
|
complement component 5
|
|
chr20_+_123230
|
0.193
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr3_+_51895644
|
0.188
|
NM_203424
|
IQCF2
|
IQ motif containing F2
|
|
chr2_-_224467142
|
0.182
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr1_+_145096391
|
0.180
|
NM_004892
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chr12_-_10588543
|
0.180
|
NM_002260
|
KLRC2
KLRC3
|
killer cell lectin-like receptor subfamily C, member 2
killer cell lectin-like receptor subfamily C, member 3
|
|
chr10_+_104613966
|
0.177
|
NM_001136200
NM_144591
|
C10orf32-AS3MT
C10orf32
|
C10orf32-AS3MT readthrough
chromosome 10 open reading frame 32
|
|
chr14_-_20774089
|
0.176
|
|
TTC5
|
tetratricopeptide repeat domain 5
|
|
chr15_-_31947541
|
0.175
|
NM_130901
|
OTUD7A
|
OTU domain containing 7A
|
|
chr15_-_89089792
|
0.174
|
NM_001144074
NM_017996
|
DET1
|
de-etiolated homolog 1 (Arabidopsis)
|
|
chr6_+_73076431
|
0.171
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr3_-_49851217
|
0.170
|
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr14_-_20774129
|
0.169
|
NM_138376
|
TTC5
|
tetratricopeptide repeat domain 5
|
|
chr3_-_100712248
|
0.166
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr5_-_39394423
|
0.165
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr2_+_210444154
|
0.165
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr19_-_48752921
|
0.164
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr6_-_117747017
|
0.164
|
NM_002944
|
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase
|
|
chr1_+_171060017
|
0.163
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr1_-_89591685
|
0.160
|
NM_004120
|
GBP2
|
guanylate binding protein 2, interferon-inducible
|
|
chr4_+_141264596
|
0.160
|
NM_001153446
NM_001153635
NM_001153585
NM_001153690
|
SCOC
|
short coiled-coil protein
|
|
chr10_+_69869249
|
0.158
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr8_-_108510094
|
0.158
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr1_+_196946666
|
0.156
|
NM_030787
|
CFHR5
|
complement factor H-related 5
|
|
chr2_+_109204745
|
0.156
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr13_+_46276445
|
0.155
|
NM_152719
|
SPERT
|
spermatid associated
|
|
chr6_-_33041266
|
0.155
|
NM_033554
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1
|
|
chr2_+_152214105
|
0.155
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr5_+_54320083
|
0.154
|
NM_002104
|
GZMK
|
granzyme K (granzyme 3; tryptase II)
|
|
chr3_+_113616317
|
0.153
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr10_+_111765725
|
0.153
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr1_+_196788860
|
0.151
|
NM_002113
|
CFHR1
|
complement factor H-related 1
|
|
chr2_+_109204904
|
0.147
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr3_-_138478184
|
0.146
|
NM_006219
|
PIK3CB
|
phosphoinositide-3-kinase, catalytic, beta polypeptide
|
|
chr3_-_164796275
|
0.145
|
NM_001041
|
SI
|
sucrase-isomaltase (alpha-glucosidase)
|
|
chr17_-_67138013
|
0.144
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr2_-_171923482
|
0.144
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr5_+_140602914
|
0.143
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr16_+_55515468
|
0.140
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chrX_-_57164057
|
0.139
|
NM_019003
|
SPIN2A
|
spindlin family, member 2A
|
|
chrX_-_19988381
|
0.139
|
NM_198279
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr6_+_46761093
|
0.136
|
NM_005588
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase)
|
|
chr13_-_49110117
|
0.134
|
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2
|
|
chr15_-_93632161
|
0.132
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr1_+_160336856
|
0.132
|
NM_005598
|
NHLH1
|
nescient helix loop helix 1
|
|
chr10_+_89117424
|
0.132
|
NM_001009610
|
FAM22A
FAM22D
|
family with sequence similarity 22, member A
family with sequence similarity 22, member D
|
|
chr5_-_147286064
|
0.130
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr8_+_27182995
|
0.129
|
NM_173176
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr3_-_195310985
|
0.129
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr2_-_224467095
|
0.128
|
|
SCG2
|
secretogranin II
|
|
chr17_-_38978824
|
0.126
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr1_-_1711453
|
0.125
|
NM_001198994
|
NADK
|
NAD kinase
|
|
chr17_+_3377346
|
0.125
|
NM_001128085
|
ASPA
|
aspartoacylase
|
|
chr4_-_186697065
|
0.125
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chrX_+_135278912
|
0.124
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr2_+_79347487
|
0.124
|
NM_002909
|
REG1A
|
regenerating islet-derived 1 alpha
|
|
chr15_+_25101697
|
0.124
|
NM_022805
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr9_-_95244634
|
0.121
|
|
ASPN
|
asporin
|
|
chr9_+_79074067
|
0.119
|
NM_001097633
NM_001490
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chrX_+_22050920
|
0.119
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr12_+_72332625
|
0.118
|
NM_173353
|
TPH2
|
tryptophan hydroxylase 2
|
|
chrX_-_8139307
|
0.118
|
NM_016378
|
VCX2
|
variable charge, X-linked 2
|
|
chr17_-_39222126
|
0.116
|
NM_033184
|
KRTAP2-4
|
keratin associated protein 2-4
|
|
chr6_+_16143281
|
0.115
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr2_+_79347610
|
0.115
|
|
REG1A
|
regenerating islet-derived 1 alpha
|
|
chr5_+_137203559
|
0.114
|
|
MYOT
|
myotilin
|
|
chr17_+_33448597
|
0.113
|
NM_017559
|
FNDC8
|
fibronectin type III domain containing 8
|
|
chr3_+_46395224
|
0.112
|
NM_001123041
NM_001123396
|
CCR2
|
chemokine (C-C motif) receptor 2
|
|
chr3_-_151102536
|
0.112
|
NM_022788
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12
|
|
chr3_-_169587598
|
0.111
|
NM_024727
|
LRRC31
|
leucine rich repeat containing 31
|
|
chr10_+_124670216
|
0.111
|
NM_001029888
|
FAM24A
|
family with sequence similarity 24, member A
|
|
chr4_-_156787377
|
0.110
|
NM_017419
|
ACCN5
|
amiloride-sensitive cation channel 5, intestinal
|
|
chrX_-_99987072
|
0.109
|
NM_001129896
|
SYTL4
|
synaptotagmin-like 4
|
|
chr1_+_101185195
|
0.108
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr2_-_79315118
|
0.106
|
NM_006507
|
REG1B
|
regenerating islet-derived 1 beta
|
|
chr7_-_14880954
|
0.105
|
NM_004080
NM_145695
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chr1_-_201465700
|
0.104
|
NM_001193572
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr6_+_20534686
|
0.103
|
NM_017774
|
CDKAL1
|
CDK5 regulatory subunit associated protein 1-like 1
|
|
chr11_-_18956545
|
0.101
|
NM_147199
|
MRGPRX1
|
MAS-related GPR, member X1
|
|
chr3_-_114790221
|
0.101
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr1_-_93399531
|
0.101
|
|
|
|
|
chr17_+_78356710
|
0.100
|
|
RNF213
|
ring finger protein 213
|
|
chr2_-_175712276
|
0.100
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr8_-_42623928
|
0.099
|
NM_001199279
NM_004198
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6
|
|
chr3_+_112051914
|
0.099
|
NM_001004196
NM_005944
|
CD200
|
CD200 molecule
|
|
chr12_-_50790391
|
0.097
|
NM_001145475
|
FAM186A
|
family with sequence similarity 186, member A
|
|
chr10_+_88985204
|
0.097
|
NM_001099338
|
FAM22A
FAM22D
|
family with sequence similarity 22, member A
family with sequence similarity 22, member D
|
|
chr4_-_170679027
|
0.096
|
NM_017867
|
C4orf27
|
chromosome 4 open reading frame 27
|
|
chr20_+_31870940
|
0.096
|
NM_033197
|
BPIFB1
|
BPI fold containing family B, member 1
|
|
chr13_+_49684388
|
0.095
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr2_+_36923832
|
0.094
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr9_+_96846710
|
0.093
|
NM_001253829
NM_152422
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chr8_+_27183047
|
0.092
|
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr17_+_18647325
|
0.092
|
NM_031456
|
FBXW10
|
F-box and WD repeat domain containing 10
|
|
chr17_+_3379295
|
0.092
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr6_+_72926144
|
0.089
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chrX_+_154255063
|
0.089
|
NM_023934
|
FUNDC2
|
FUN14 domain containing 2
|
|
chr9_-_21077804
|
0.089
|
NM_002176
|
IFNB1
|
interferon, beta 1, fibroblast
|
|
chr1_+_196912910
|
0.089
|
NM_005666
|
CFHR2
|
complement factor H-related 2
|
|
chrX_+_108779009
|
0.088
|
NM_018698
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr11_+_59807747
|
0.088
|
NM_173801
|
PLAC1L
|
placenta-specific 1-like
|
|
chr3_-_160101443
|
0.087
|
NM_001190242
|
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas)
|
|
chr4_-_140060601
|
0.086
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr18_-_40695483
|
0.085
|
NM_002930
|
RIT2
|
Ras-like without CAAX 2
|
|
chrX_+_107683015
|
0.085
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr1_+_162602308
|
0.085
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr5_+_141346401
|
0.085
|
NM_001201365
NM_183399
|
RNF14
|
ring finger protein 14
|
|
chr18_+_54424444
|
0.084
|
|
WDR7
|
WD repeat domain 7
|
|
chr5_-_58295758
|
0.084
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_-_159092509
|
0.084
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr4_+_71263598
|
0.084
|
NM_021225
|
PROL1
|
proline rich, lacrimal 1
|
|
chr14_+_60712469
|
0.083
|
NM_177952
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr20_+_2276612
|
0.082
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr17_-_29641068
|
0.082
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr12_+_60083125
|
0.082
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr3_+_180320955
|
0.081
|
|
TTC14
|
tetratricopeptide repeat domain 14
|
|
chr9_-_21187529
|
0.081
|
NM_021068
|
IFNA4
|
interferon, alpha 4
|
|
chr12_+_8325149
|
0.080
|
NM_001004328
|
ZNF705A
|
zinc finger protein 705A
|
|
chr7_+_149570004
|
0.079
|
NM_001100592
NM_145230
|
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2
|
|
chr8_-_7220489
|
0.077
|
NM_001164457
|
ZNF705G
|
zinc finger protein 705G
|
|
chr6_+_28109611
|
0.076
|
NM_006298
|
ZNF192
|
zinc finger protein 192
|
|
chr1_+_162466963
|
0.076
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr2_-_122159140
|
0.075
|
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr5_-_59783885
|
0.075
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr18_-_52625278
|
0.075
|
NM_001143829
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr17_-_39165343
|
0.075
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chrX_-_49460568
|
0.073
|
NM_003785
|
PAGE1
|
P antigen family, member 1 (prostate associated)
|
|
chr4_-_118006731
|
0.073
|
NM_152402
|
TRAM1L1
|
translocation associated membrane protein 1-like 1
|
|
chr7_+_29519485
|
0.072
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr18_-_29340698
|
0.072
|
NM_001034172
|
MCART2
|
mitochondrial carrier triple repeat 2
|
|
chr10_-_50342052
|
0.070
|
NM_001164484
|
FAM170B
|
family with sequence similarity 170, member B
|
|
chr11_-_40315663
|
0.069
|
NM_020929
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr4_-_70080448
|
0.068
|
NM_001073
|
UGT2B11
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B11
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr6_+_47749774
|
0.068
|
NM_181744
|
OPN5
|
opsin 5
|
|
chr5_+_140743755
|
0.068
|
NM_018918
NM_032054
|
PCDHGA5
|
protocadherin gamma subfamily A, 5
|
|
chr5_+_71502700
|
0.068
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr5_+_140729723
|
0.068
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr1_-_101360405
|
0.067
|
NM_001033025
NM_001439
|
EXTL2
|
exostoses (multiple)-like 2
|
|
chr11_+_67376926
|
0.067
|
|
NDUFV1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa
|
|
chr5_+_137673223
|
0.067
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr4_-_104020964
|
0.067
|
NM_020139
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2
|
|
chr6_-_52628272
|
0.066
|
NM_000846
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chr2_+_54342409
|
0.065
|
NM_138448
|
ACYP2
|
acylphosphatase 2, muscle type
|
|
chr2_-_128439359
|
0.065
|
NM_001161404
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr10_+_15085894
|
0.065
|
NM_001039702
NM_018324
|
OLAH
|
oleoyl-ACP hydrolase
|
|
chr4_+_100495972
|
0.065
|
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr1_-_153283166
|
0.065
|
NM_052891
|
PGLYRP3
|
peptidoglycan recognition protein 3
|
|
chr20_+_56964162
|
0.064
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr20_+_57470200
|
0.064
|
|
GNAS
|
GNAS complex locus
|
|
chr6_-_41168923
|
0.063
|
NM_024807
|
TREML2
|
triggering receptor expressed on myeloid cells-like 2
|
|
chr9_-_19378815
|
0.063
|
|
RPS6
|
ribosomal protein S6
|
|
chr4_+_88742549
|
0.063
|
NM_001184694
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr5_+_57787260
|
0.062
|
NM_152687
|
GAPT
|
GRB2-binding adaptor protein, transmembrane
|
|
chr5_+_59784004
|
0.062
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chrX_-_102774416
|
0.062
|
NM_080879
|
RAB40A
|
RAB40A, member RAS oncogene family
|
|
chr20_+_138121
|
0.062
|
NM_139074
|
DEFB127
|
defensin, beta 127
|
|
chr9_-_91793681
|
0.061
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr2_+_233498206
|
0.061
|
NM_025202
|
EFHD1
|
EF-hand domain family, member D1
|
|
chr12_+_7167979
|
0.060
|
NM_001734
NM_201442
|
C1S
|
complement component 1, s subcomponent
|