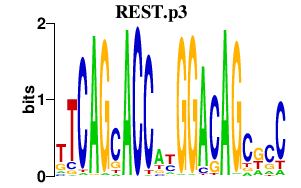
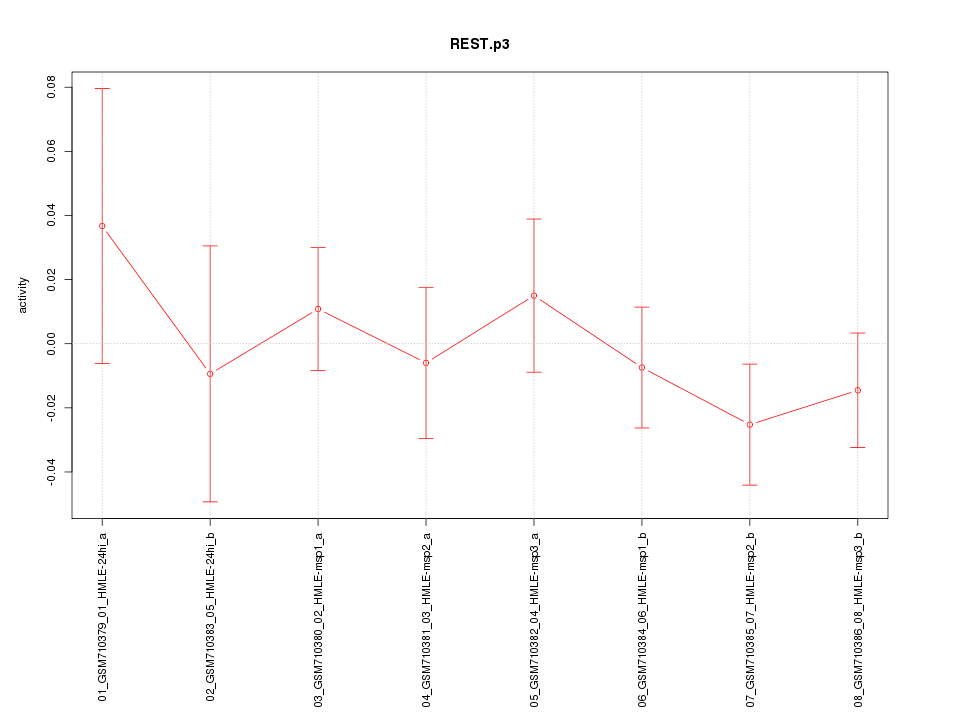
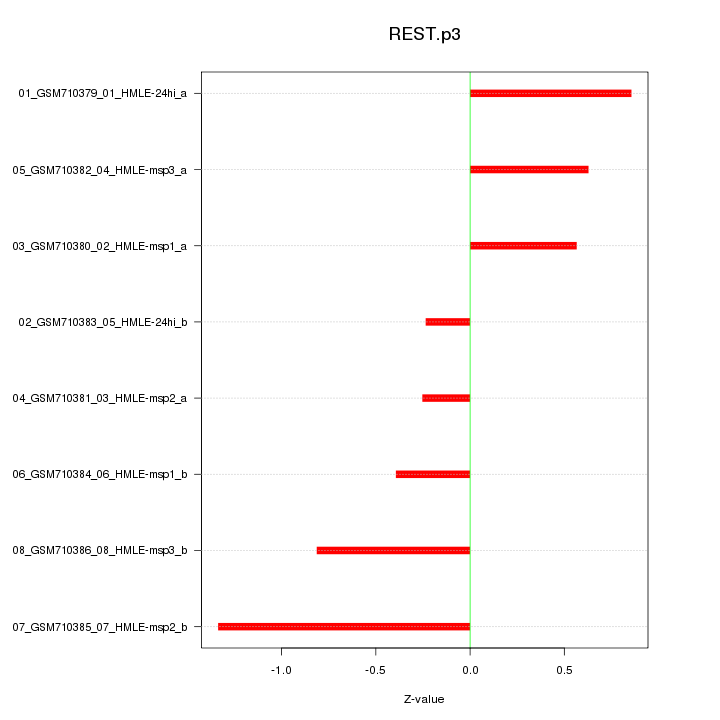
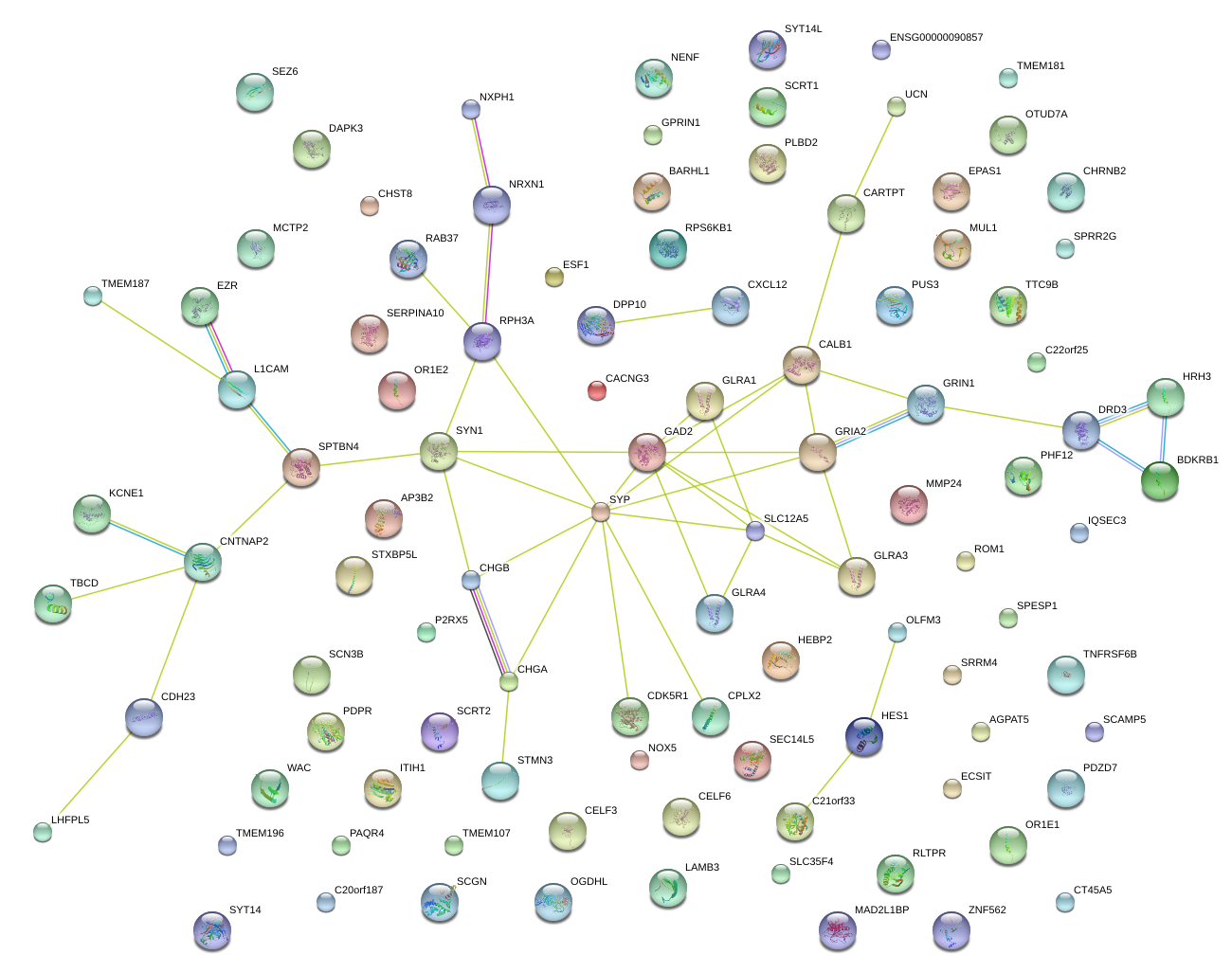
|
chrX_-_153141398
|
0.955
|
NM_000425
NM_001143963
NM_024003
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr15_-_72612247
|
0.451
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr17_+_30813928
|
0.449
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr15_-_72612497
|
0.448
|
NM_052840
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr3_+_193853937
|
0.424
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr3_+_193853930
|
0.419
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr8_-_91094924
|
0.393
|
NM_004929
|
CALB1
|
calbindin 1, 28kDa
|
|
chr7_+_145813452
|
0.369
|
NM_014141
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr10_-_50970321
|
0.340
|
NM_001143996
NM_001143997
NM_018245
|
OGDHL
|
oxoglutarate dehydrogenase-like
|
|
chr20_+_62328068
|
0.327
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr8_-_145559831
|
0.284
|
NM_031309
|
SCRT1
|
scratch homolog 1, zinc finger protein (Drosophila)
|
|
chr20_+_62328002
|
0.284
|
NM_003823
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr11_-_123524881
|
0.277
|
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr2_+_46524448
|
0.262
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr17_-_3301703
|
0.238
|
NM_003553
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1
|
|
chr17_-_3337134
|
0.233
|
NM_003554
|
OR1E2
|
olfactory receptor, family 1, subfamily E, member 2
|
|
chrX_-_153141284
|
0.230
|
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr20_-_62284701
|
0.220
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr9_+_135457974
|
0.199
|
NM_020064
|
BARHL1
|
BarH-like homeobox 1
|
|
chr14_+_93389491
|
0.187
|
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr15_-_72612127
|
0.167
|
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chrX_-_49056638
|
0.162
|
NM_003179
|
SYP
|
synaptophysin
|
|
chr15_+_75287829
|
0.149
|
NM_001178111
NM_001178112
NM_138967
|
SCAMP5
|
secretory carrier membrane protein 5
|
|
chr1_+_154540250
|
0.149
|
NM_000748
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal)
|
|
chr1_+_210111575
|
0.133
|
|
SYT14
|
synaptotagmin XIV
|
|
chr17_+_72667255
|
0.127
|
NM_175738
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr3_+_120627026
|
0.123
|
NM_014980
|
STXBP5L
|
syntaxin binding protein 5-like
|
|
chr12_+_113796374
|
0.122
|
|
PLBD2
|
phospholipase B domain containing 2
|
|
chr11_-_125773086
|
0.118
|
NM_031307
|
PUS3
|
pseudouridylate synthase 3
|
|
chr8_+_57358361
|
0.117
|
|
|
|
|
chr20_+_44657858
|
0.116
|
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr20_+_5891973
|
0.116
|
NM_001819
|
CHGB
|
chromogranin B (secretogranin 1)
|
|
chr19_+_34175433
|
0.113
|
NM_022467
|
CHST8
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 8
|
|
chr1_-_209825797
|
0.108
|
NM_000228
|
LAMB3
|
laminin, beta 3
|
|
chr12_+_175872
|
0.108
|
NM_001170738
|
IQSEC3
|
IQ motif and Sec7 domain 3
|
|
chr14_+_93389444
|
0.101
|
NM_001275
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr11_-_1769086
|
0.101
|
|
IFITM10
|
interferon induced transmembrane protein 10
|
|
chr19_-_3971089
|
0.100
|
|
DAPK3
|
death-associated protein kinase 3
|
|
chr1_+_210111537
|
0.098
|
NM_001146261
NM_001146262
NM_001146264
NM_153262
|
SYT14
|
synaptotagmin XIV
|
|
chr16_+_3019341
|
0.097
|
NM_152341
|
PAQR4
|
progestin and adipoQ receptor family member IV
|
|
chrX_-_47479229
|
0.097
|
NM_006950
NM_133499
|
SYN1
|
synapsin I
|
|
chr12_+_113796370
|
0.092
|
NM_001159727
NM_173542
|
PLBD2
|
phospholipase B domain containing 2
|
|
chr2_+_115199898
|
0.091
|
NM_020868
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr19_-_3971032
|
0.090
|
|
DAPK3
|
death-associated protein kinase 3
|
|
chr8_+_6566153
|
0.087
|
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr5_+_175298492
|
0.085
|
NM_001008220
|
CPLX2
|
complexin 2
|
|
chr20_-_13765509
|
0.083
|
NM_016649
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae)
|
|
chr7_+_8473584
|
0.078
|
NM_152745
|
NXPH1
|
neurexophilin 1
|
|
chr7_+_8473237
|
0.077
|
|
NXPH1
|
neurexophilin 1
|
|
chr17_-_3599487
|
0.076
|
NM_001204519
NM_001204520
NM_002561
NM_175080
|
P2RX5-TAX1BP3
P2RX5
|
P2RX5-TAX1BP3 readthrough
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr18_-_5197240
|
0.075
|
NM_001145194
|
C18orf42
|
chromosome 18 open reading frame 42
|
|
chr5_+_175298604
|
0.073
|
|
CPLX2
|
complexin 2
|
|
chr1_-_102462789
|
0.073
|
NM_058170
|
OLFM3
|
olfactomedin 3
|
|
chr20_-_60795270
|
0.072
|
NM_007232
|
HRH3
|
histamine receptor H3
|
|
chr5_+_175298538
|
0.072
|
|
CPLX2
|
complexin 2
|
|
chr16_+_67686813
|
0.071
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr8_+_6565870
|
0.067
|
NM_018361
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr20_+_33814538
|
0.067
|
NM_006690
|
MMP24
|
matrix metallopeptidase 24 (membrane-inserted)
|
|
chr2_-_51259239
|
0.065
|
|
NRXN1
|
neurexin 1
|
|
chr14_-_74892804
|
0.065
|
NM_001105579
|
SYNDIG1L
|
synapse differentiation inducing 1-like
|
|
chr6_+_138725314
|
0.062
|
NM_014320
|
HEBP2
|
heme binding protein 2
|
|
chr16_+_70147528
|
0.062
|
NM_017990
|
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit
|
|
chr7_+_8474016
|
0.062
|
|
NXPH1
|
neurexophilin 1
|
|
chr6_+_35773070
|
0.061
|
NM_182548
|
LHFPL5
|
lipoma HMGIC fusion partner-like 5
|
|
chr16_+_24267474
|
0.060
|
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3
|
|
chr5_-_151304322
|
0.055
|
NM_000171
NM_001146040
|
GLRA1
|
glycine receptor, alpha 1
|
|
chr7_-_19812403
|
0.055
|
NM_152774
|
TMEM196
|
transmembrane protein 196
|
|
chr6_+_158957467
|
0.054
|
NM_020823
|
TMEM181
|
transmembrane protein 181
|
|
chr3_-_113897849
|
0.051
|
NM_000796
NM_033663
|
DRD3
|
dopamine receptor D3
|
|
chr9_+_140033588
|
0.049
|
NM_000832
NM_001185090
NM_001185091
NM_007327
NM_021569
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr1_-_102462371
|
0.048
|
|
OLFM3
|
olfactomedin 3
|
|
chr5_+_71014989
|
0.047
|
NM_004291
|
CARTPT
|
CART prepropeptide
|
|
chr1_-_151689246
|
0.046
|
NM_007185
|
CELF3
|
CUGBP, Elav-like family member 3
|
|
chr10_+_73555527
|
0.044
|
NM_001171933
NM_001171934
|
CDH23
|
cadherin-related 23
|
|
chr11_+_62380212
|
0.042
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chr6_+_138725458
|
0.041
|
|
HEBP2
|
heme binding protein 2
|
|
chr10_-_44880452
|
0.040
|
NM_000609
NM_001033886
NM_001178134
NM_199168
|
CXCL12
|
chemokine (C-X-C motif) ligand 12
|
|
chr17_-_27332888
|
0.040
|
NM_001098635
NM_178860
|
SEZ6
|
seizure related 6 homolog (mouse)
|
|
chr19_-_40724272
|
0.040
|
NM_152479
|
TTC9B
|
tetratricopeptide repeat domain 9B
|
|
chr6_+_43603554
|
0.040
|
NM_014628
|
MAD2L1BP
|
MAD2L1 binding protein
|
|
chr15_-_83378488
|
0.039
|
NM_004644
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit
|
|
chr16_+_24267672
|
0.038
|
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3
|
|
chr10_-_102790877
|
0.037
|
NM_001195263
NM_024895
|
PDZD7
|
PDZ domain containing 7
|
|
chr14_-_58332591
|
0.036
|
NM_001206920
|
SLC35F4
|
solute carrier family 35, member F4
|
|
chr4_+_158141735
|
0.034
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr2_-_27531113
|
0.033
|
NM_003353
|
UCN
|
urocortin
|
|
chr1_+_212606194
|
0.033
|
NM_013349
|
NENF
|
neudesin neurotrophic factor
|
|
chr3_+_52812438
|
0.032
|
NM_001166434
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr10_-_102790850
|
0.032
|
|
PDZD7
|
PDZ domain containing 7
|
|
chr10_+_26505593
|
0.032
|
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa)
|
|
chr19_+_41036367
|
0.031
|
NM_025213
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr15_+_69222838
|
0.029
|
NM_145658
NM_001184780
|
SPESP1
NOX5
|
sperm equatorial segment protein 1
NADPH oxidase, EF-hand calcium binding domain 5
|
|
chr6_+_158957652
|
0.029
|
|
TMEM181
|
transmembrane protein 181
|
|
chr1_-_20834609
|
0.028
|
NM_024544
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1
|
|
chr16_+_5008317
|
0.026
|
NM_014692
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae)
|
|
chr21_-_35828038
|
0.026
|
NM_001127670
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr15_-_32162778
|
0.025
|
|
OTUD7A
|
OTU domain containing 7A
|
|
chr19_-_11639929
|
0.025
|
NM_001142464
NM_001142465
NM_016581
|
ECSIT
|
ECSIT homolog (Drosophila)
|
|
chr1_-_153123426
|
0.024
|
NM_001014291
|
SPRR2G
|
small proline-rich protein 2G
|
|
chr6_+_43603588
|
0.024
|
|
MAD2L1BP
|
MAD2L1 binding protein
|
|
chr1_-_20834516
|
0.023
|
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1
|
|
chr4_-_175750377
|
0.022
|
NM_001042543
NM_006529
|
GLRA3
|
glycine receptor, alpha 3
|
|
chr19_-_11639679
|
0.020
|
NM_001243204
|
ECSIT
|
ECSIT homolog (Drosophila)
|
|
chr15_+_94774763
|
0.020
|
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chrX_+_153237990
|
0.019
|
NM_003492
|
TMEM187
|
transmembrane protein 187
|
|
chr12_+_113229548
|
0.019
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr17_-_27240982
|
0.018
|
|
PHF12
|
PHD finger protein 12
|
|
chr22_+_20008633
|
0.017
|
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr6_+_25652373
|
0.013
|
NM_006998
|
SCGN
|
secretagogin, EF-hand calcium binding protein
|
|
chr14_-_94759594
|
0.012
|
NM_001100607
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10
|
|
chr10_+_28822428
|
0.011
|
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr12_+_119419299
|
0.010
|
NM_194286
|
SRRM4
|
serine/arginine repetitive matrix 4
|
|
chr2_-_51259536
|
0.010
|
NM_001135659
NM_004801
|
NRXN1
|
neurexin 1
|
|
chr19_-_45737466
|
0.009
|
NM_138568
|
EXOC3L2
|
exocyst complex component 3-like 2
|
|
chr11_-_128807712
|
0.009
|
NM_001195194
|
TP53AIP1
|
tumor protein p53 regulated apoptosis inducing protein 1
|
|
chr10_+_28822387
|
0.009
|
NM_016628
NM_100486
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr4_+_8582216
|
0.008
|
NM_080819
|
GPR78
|
G protein-coupled receptor 78
|
|
chr19_-_55691468
|
0.008
|
NM_003180
|
SYT5
|
synaptotagmin V
|
|
chr5_+_141303366
|
0.008
|
NM_001142603
NM_014773
|
KIAA0141
|
KIAA0141
|
|
chr17_-_65362635
|
0.006
|
|
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12
|
|
chr8_+_68864341
|
0.006
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr1_+_171750760
|
0.005
|
NM_001007239
NM_014955
NM_015935
|
METTL13
|
methyltransferase like 13
|
|
chr19_+_15197790
|
0.004
|
NM_001004713
|
OR1I1
|
olfactory receptor, family 1, subfamily I, member 1
|
|
chr11_+_32112607
|
0.004
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr1_+_171750829
|
0.003
|
|
METTL13
|
methyltransferase like 13
|
|
chr2_+_131513782
|
0.003
|
NM_001105194
NM_001105195
|
FAM123C
|
family with sequence similarity 123C
|
|
chr7_+_134832765
|
0.003
|
NM_018295
|
TMEM140
|
transmembrane protein 140
|
|
chr6_-_71666740
|
0.003
|
NM_080742
|
B3GAT2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S)
|
|
chr1_+_171750807
|
0.002
|
|
METTL13
|
methyltransferase like 13
|
|
chr12_-_99288502
|
0.002
|
NM_001204065
NM_001204079
NM_001204080
NM_001204081
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr22_+_31795538
|
0.001
|
NM_004147
|
DRG1
|
developmentally regulated GTP binding protein 1
|
|
chr5_+_69696805
|
0.001
|
|
SMA4
|
glucuronidase, beta pseudogene
|
|
chr11_+_637268
|
0.001
|
NM_000797
|
DRD4
|
dopamine receptor D4
|
|
chr9_-_74061819
|
0.001
|
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr17_-_65362667
|
0.000
|
NM_002816
NM_174871
|
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12
|