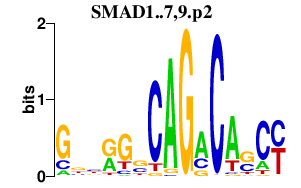
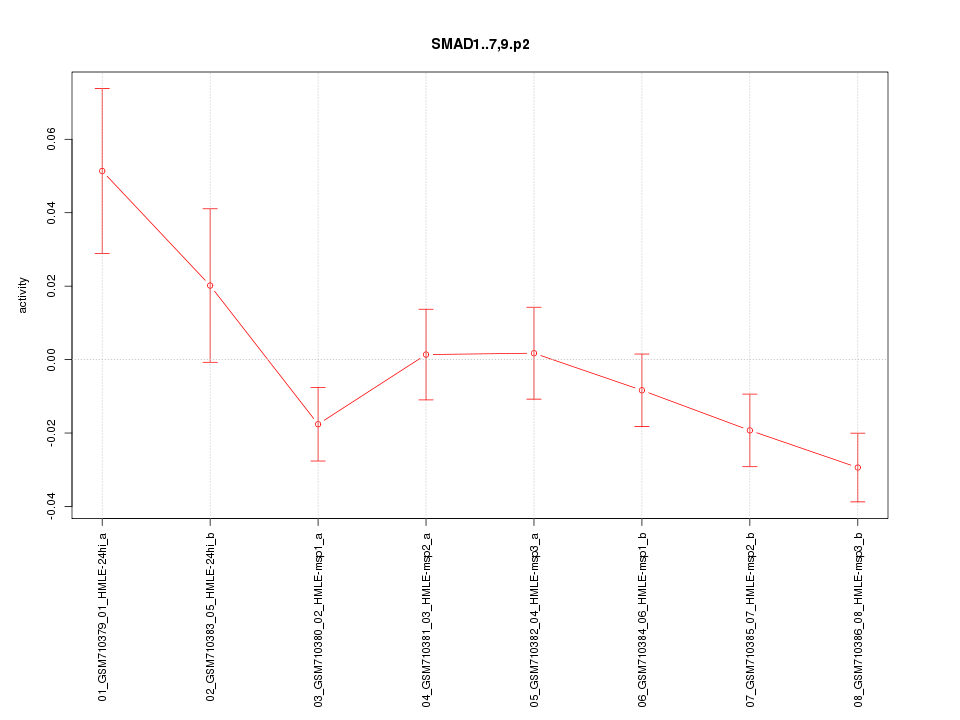
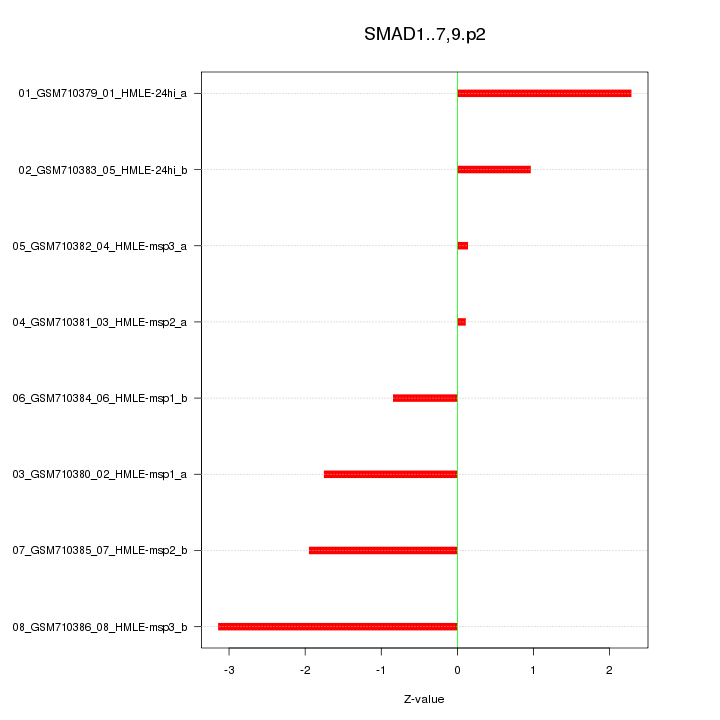
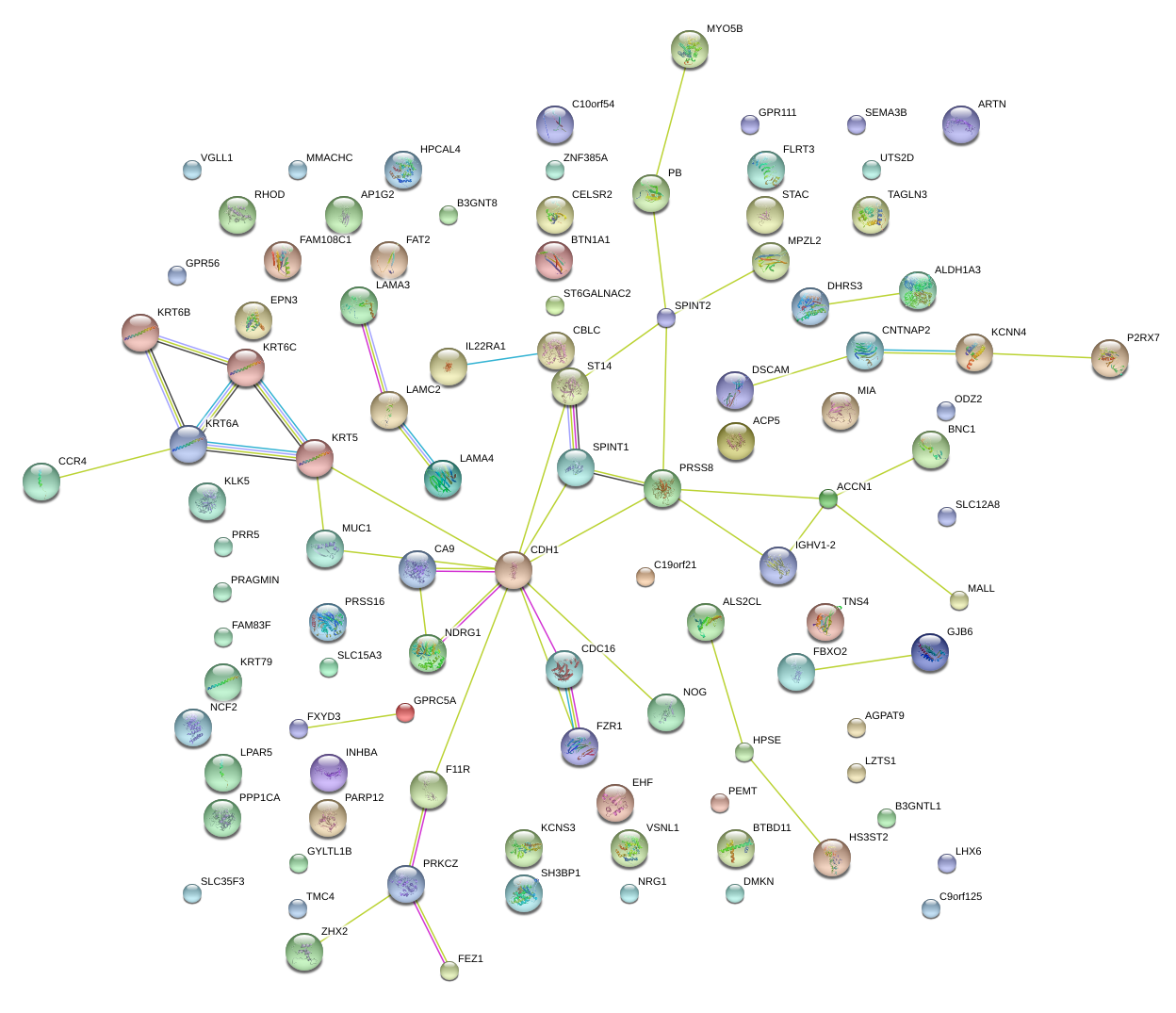
|
chr19_+_35607218
|
2.628
|
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr19_-_35992798
|
2.100
|
NM_001035516
|
DMKN
|
dermokine
|
|
chr8_+_32504250
|
2.075
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr19_+_751108
|
1.797
|
NM_173481
|
C19orf21
|
chromosome 19 open reading frame 21
|
|
chr22_+_38035683
|
1.747
|
NM_018957
|
SH3BP1
|
SH3-domain binding protein 1
|
|
chr1_+_15272414
|
1.641
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr16_+_57662301
|
1.621
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr15_+_41136178
|
1.588
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr1_-_160990862
|
1.561
|
|
F11R
|
F11 receptor
|
|
chr1_-_160990971
|
1.543
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr11_+_66824308
|
1.464
|
|
RHOD
|
ras homolog gene family, member D
|
|
chr16_+_22825481
|
1.454
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr12_-_52867521
|
1.444
|
NM_173086
|
KRT6C
|
keratin 6C
|
|
chr1_+_183155173
|
1.441
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr18_-_47376156
|
1.403
|
|
MYO5B
|
myosin VB
|
|
chr2_-_110874142
|
1.389
|
NM_005434
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr22_+_40390935
|
1.360
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr1_+_109792640
|
1.356
|
NM_001408
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr11_+_130029663
|
1.356
|
NM_021978
|
ST14
|
suppression of tumorigenicity 14 (colon carcinoma)
|
|
chr8_-_8239188
|
1.348
|
NM_001080826
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr1_+_183155393
|
1.337
|
|
LAMC2
|
laminin, gamma 2
|
|
chr11_+_130029699
|
1.329
|
|
ST14
|
suppression of tumorigenicity 14 (colon carcinoma)
|
|
chr11_+_130029712
|
1.309
|
|
ST14
|
suppression of tumorigenicity 14 (colon carcinoma)
|
|
chr1_+_183155361
|
1.305
|
|
LAMC2
|
laminin, gamma 2
|
|
chr15_+_41136622
|
1.283
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr11_-_118134956
|
1.259
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr11_-_118135197
|
1.250
|
NM_005797
NM_144765
|
MPZL2
|
myelin protein zero-like 2
|
|
chr2_+_17721806
|
1.235
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr1_+_234349995
|
1.166
|
|
SLC35F3
|
solute carrier family 35, member F3
|
|
chr19_-_54676813
|
1.102
|
NM_001145303
NM_144686
|
TMC4
|
transmembrane channel-like 4
|
|
chr1_+_44398991
|
1.089
|
NM_001136215
NM_057090
NM_057091
|
ARTN
|
artemin
|
|
chr16_-_31147062
|
1.050
|
NM_002773
|
PRSS8
|
protease, serine, 8
|
|
chr11_-_125366048
|
1.045
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr11_-_118134986
|
1.029
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr7_-_41742666
|
0.995
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr2_+_17721983
|
0.988
|
|
VSNL1
|
visinin-like 1
|
|
chr8_-_134309448
|
0.982
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr19_-_41933255
|
0.966
|
|
|
|
|
chrX_+_135614310
|
0.954
|
NM_016267
|
VGLL1
|
vestigial like 1 (Drosophila)
|
|
chr15_-_83953465
|
0.937
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr2_-_110873638
|
0.933
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr12_-_6740814
|
0.918
|
NM_001142961
|
LPAR5
|
lysophosphatidic acid receptor 5
|
|
chr15_+_101420004
|
0.910
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr2_+_18059923
|
0.908
|
NM_002252
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3
|
|
chr8_+_123793894
|
0.881
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr16_+_68771214
|
0.878
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr1_+_2005424
|
0.876
|
NM_001242874
|
PRKCZ
|
protein kinase C, zeta
|
|
chr5_+_166711842
|
0.863
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr9_-_124984018
|
0.862
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr16_+_68771192
|
0.856
|
NM_004360
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr19_-_51456159
|
0.855
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr18_+_21452981
|
0.853
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr12_+_107712151
|
0.852
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr9_+_35673852
|
0.840
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr6_+_27215479
|
0.838
|
NM_005865
|
PRSS16
|
protease, serine, 16 (thymus)
|
|
chr3_+_36421944
|
0.828
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr7_+_148036528
|
0.828
|
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr6_+_26501448
|
0.819
|
NM_001732
|
BTN1A1
|
butyrophilin, subfamily 1, member A1
|
|
chr3_-_191048324
|
0.806
|
NM_198152
|
UTS2D
|
urotensin 2 domain containing
|
|
chr16_-_31146992
|
0.806
|
|
PRSS8
|
protease, serine, 8
|
|
chr12_-_52914080
|
0.799
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr19_+_38755097
|
0.798
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr13_-_20806533
|
0.789
|
NM_001110219
NM_001110220
NM_001110221
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr8_+_123794043
|
0.784
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr17_+_54671059
|
0.776
|
NM_005450
|
NOG
|
noggin
|
|
chr17_-_74581934
|
0.774
|
|
ST6GALNAC2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2
|
|
chr17_-_38657814
|
0.771
|
NM_032865
|
TNS4
|
tensin 4
|
|
chr3_-_124930192
|
0.757
|
NM_001195483
|
SLC12A8
|
solute carrier family 12 (potassium/chloride transporters), member 8
|
|
chr14_-_24037266
|
0.748
|
NM_003917
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr21_-_42219038
|
0.733
|
NM_001389
|
DSCAM
|
Down syndrome cell adhesion molecule
|
|
chr4_-_84256304
|
0.731
|
NM_001166498
NM_006665
|
HPSE
|
heparanase
|
|
chr19_+_41281281
|
0.729
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr10_-_73533196
|
0.717
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr1_-_11714471
|
0.712
|
|
FBXO2
|
F-box protein 2
|
|
chr19_+_45281125
|
0.709
|
NM_001130852
NM_012116
|
CBLC
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence c
|
|
chr3_+_111717585
|
0.705
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr12_+_13043955
|
0.704
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr11_+_45944155
|
0.697
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr19_-_11689591
|
0.691
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr3_-_46718898
|
0.687
|
NM_182775
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr17_+_48610045
|
0.683
|
NM_017957
|
EPN3
|
epsin 3
|
|
chr11_+_34654010
|
0.681
|
NM_001206616
|
EHF
|
ets homologous factor
|
|
chr15_+_101420032
|
0.680
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr1_-_12677347
|
0.673
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr15_+_80987609
|
0.669
|
NM_021214
|
FAM108C1
|
family with sequence similarity 108, member C1
|
|
chr9_-_104249333
|
0.667
|
NM_032342
|
C9orf125
|
chromosome 9 open reading frame 125
|
|
chr4_+_84457390
|
0.661
|
NM_032717
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr7_-_139763520
|
0.657
|
NM_022750
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr19_-_41934634
|
0.655
|
NM_198540
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8
|
|
chr16_+_68771304
|
0.653
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr11_+_66824285
|
0.643
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr12_-_54778456
|
0.642
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr1_-_11714396
|
0.642
|
|
FBXO2
|
F-box protein 2
|
|
chr22_+_45072936
|
0.638
|
|
PRR5
|
proline rich 5 (renal)
|
|
chr6_+_47624222
|
0.638
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chr11_-_60719116
|
0.635
|
NM_016582
|
SLC15A3
|
solute carrier family 15, member 3
|
|
chr20_-_14318200
|
0.633
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr19_+_41281081
|
0.621
|
NM_001202553
|
MIA
|
melanoma inhibitory activity
|
|
chr3_+_50304989
|
0.614
|
NM_001005914
NM_004636
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr17_-_17480325
|
0.609
|
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr1_-_183559463
|
0.609
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr1_-_24469571
|
0.597
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr19_-_44285012
|
0.581
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr9_-_99801365
|
0.579
|
NM_001333
|
CTSL2
|
cathepsin L2
|
|
chr12_-_53343607
|
0.578
|
|
KRT8
|
keratin 8
|
|
chr1_-_204329043
|
0.568
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr6_-_30043547
|
0.562
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr7_-_98030426
|
0.559
|
NM_018842
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
chr5_+_131409484
|
0.557
|
NM_000758
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage)
|
|
chr1_-_10856704
|
0.550
|
NM_001079843
NM_017766
|
CASZ1
|
castor zinc finger 1
|
|
chr17_+_73717515
|
0.541
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr15_+_33603162
|
0.541
|
NM_001036
NM_001243996
|
RYR3
|
ryanodine receptor 3
|
|
chr14_-_61748510
|
0.540
|
NM_001017970
|
TMEM30B
|
transmembrane protein 30B
|
|
chr3_-_171177851
|
0.536
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr22_-_39640956
|
0.534
|
NM_002608
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr7_-_36429695
|
0.531
|
NM_001100425
|
KIAA0895
|
KIAA0895
|
|
chr15_-_72598655
|
0.526
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr3_+_112931374
|
0.526
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr19_+_7702020
|
0.520
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr19_+_35739778
|
0.517
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr1_-_26680620
|
0.516
|
NM_001039775
|
AIM1L
|
absent in melanoma 1-like
|
|
chr20_+_62795773
|
0.513
|
|
MYT1
|
myelin transcription factor 1
|
|
chr19_-_49567123
|
0.511
|
NM_006179
|
NTF4
|
neurotrophin 4
|
|
chr6_-_138428477
|
0.511
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr14_+_65171149
|
0.500
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr16_+_82068910
|
0.499
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr19_-_11689765
|
0.498
|
NM_001111035
NM_001111036
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr19_-_51472816
|
0.497
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr18_+_21269528
|
0.497
|
NM_001127717
NM_198129
|
LAMA3
|
laminin, alpha 3
|
|
chr8_+_128748312
|
0.493
|
NM_002467
|
MYC
|
v-myc myelocytomatosis viral oncogene homolog (avian)
|
|
chr8_+_128748476
|
0.492
|
|
MYC
|
v-myc myelocytomatosis viral oncogene homolog (avian)
|
|
chr7_+_73245192
|
0.483
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr1_-_42501573
|
0.482
|
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr10_-_116392099
|
0.479
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr1_-_183559669
|
0.478
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr21_-_42218587
|
0.477
|
|
DSCAM
|
Down syndrome cell adhesion molecule
|
|
chr1_+_156123321
|
0.476
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr1_-_147245483
|
0.473
|
NM_005266
|
GJA5
|
gap junction protein, alpha 5, 40kDa
|
|
chr16_+_82068958
|
0.473
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr11_-_102668822
|
0.472
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr15_+_77287738
|
0.471
|
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chr17_+_73717556
|
0.470
|
|
ITGB4
|
integrin, beta 4
|
|
chr20_+_58515408
|
0.470
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr9_+_124329398
|
0.468
|
NM_032552
|
DAB2IP
|
DAB2 interacting protein
|
|
chr17_+_39382878
|
0.462
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr17_+_73717596
|
0.461
|
|
ITGB4
|
integrin, beta 4
|
|
chr19_+_35739519
|
0.460
|
NM_015925
NM_205834
NM_205835
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr3_+_136537860
|
0.458
|
NM_001097599
NM_001097600
NM_025246
|
TMEM22
|
transmembrane protein 22
|
|
chr12_+_47473367
|
0.458
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr11_+_1861431
|
0.457
|
NM_001145841
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr15_+_80987953
|
0.455
|
|
FAM108C1
|
family with sequence similarity 108, member C1
|
|
chr14_+_65171243
|
0.453
|
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr16_-_4466570
|
0.451
|
|
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough
coronin 7
|
|
chr19_-_44285407
|
0.438
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr1_+_16091458
|
0.436
|
NM_001024216
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr19_-_51529869
|
0.433
|
NM_001136032
|
KLK11
|
kallikrein-related peptidase 11
|
|
chr10_-_104211244
|
0.433
|
NM_024886
|
C10orf95
|
chromosome 10 open reading frame 95
|
|
chr17_+_48610077
|
0.431
|
|
EPN3
|
epsin 3
|
|
chr17_-_47841517
|
0.431
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr1_+_26872306
|
0.426
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr17_-_39092714
|
0.425
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr15_+_77287428
|
0.425
|
NM_003978
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chr3_-_69435429
|
0.424
|
NM_015123
|
FRMD4B
|
FERM domain containing 4B
|
|
chr17_-_27401590
|
0.423
|
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1
|
|
chr11_+_60869929
|
0.422
|
NM_014207
|
CD5
|
CD5 molecule
|
|
chr6_+_106534188
|
0.419
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr19_-_2038937
|
0.418
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chrX_-_84634742
|
0.418
|
NM_024921
|
POF1B
|
premature ovarian failure, 1B
|
|
chr17_-_43502992
|
0.417
|
NM_199282
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr17_-_74581881
|
0.415
|
|
ST6GALNAC2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2
|
|
chr17_-_74582144
|
0.414
|
NM_006456
|
ST6GALNAC2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2
|
|
chr1_+_4715104
|
0.412
|
NM_001042478
NM_018836
|
AJAP1
|
adherens junctions associated protein 1
|
|
chr1_+_26872342
|
0.410
|
NM_001006665
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr20_+_30598244
|
0.408
|
NM_080625
|
C20orf160
|
chromosome 20 open reading frame 160
|
|
chr1_+_158149736
|
0.404
|
NM_001766
|
CD1D
|
CD1d molecule
|
|
chr3_+_183967469
|
0.403
|
|
ECE2
|
endothelin converting enzyme 2
|
|
chrX_-_117119282
|
0.402
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr8_+_21915371
|
0.401
|
NM_001114135
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr22_+_33197678
|
0.401
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr11_+_130029773
|
0.400
|
|
ST14
|
suppression of tumorigenicity 14 (colon carcinoma)
|
|
chr3_-_50349811
|
0.400
|
NM_153281
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chrX_-_84634664
|
0.399
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr11_-_5462782
|
0.396
|
NM_001005288
|
OR51I1
|
olfactory receptor, family 51, subfamily I, member 1
|
|
chr16_-_4466595
|
0.390
|
|
CORO7
|
coronin 7
|
|
chr22_+_45072687
|
0.389
|
NM_001017530
NM_001198721
NM_015366
|
PRR5
|
proline rich 5 (renal)
|
|
chr6_+_135502445
|
0.385
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr17_-_4642638
|
0.384
|
|
CXCL16
|
chemokine (C-X-C motif) ligand 16
|
|
chr7_-_98030165
|
0.384
|
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
chr18_+_29598444
|
0.383
|
NM_017831
|
RNF125
|
ring finger protein 125
|
|
chr17_-_47841468
|
0.381
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr1_-_209979388
|
0.381
|
NM_001206696
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr11_-_119599253
|
0.381
|
NM_002855
NM_203285
NM_203286
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C)
|
|
chr3_+_183967414
|
0.380
|
NM_014693
NM_032331
|
ECE2
|
endothelin converting enzyme 2
|
|
chr17_-_27401793
|
0.375
|
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1
|
|
chr3_-_69435223
|
0.375
|
|
FRMD4B
|
FERM domain containing 4B
|
|
chr7_-_141646716
|
0.373
|
NM_013252
|
CLEC5A
|
C-type lectin domain family 5, member A
|
|
chr15_-_73661141
|
0.373
|
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chrX_-_8700170
|
0.373
|
NM_000216
|
KAL1
|
Kallmann syndrome 1 sequence
|
|
chr1_+_95558072
|
0.371
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr3_-_190040192
|
0.370
|
NM_021101
|
CLDN1
|
claudin 1
|