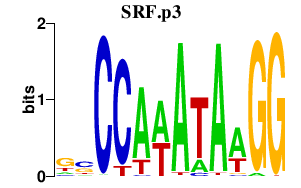
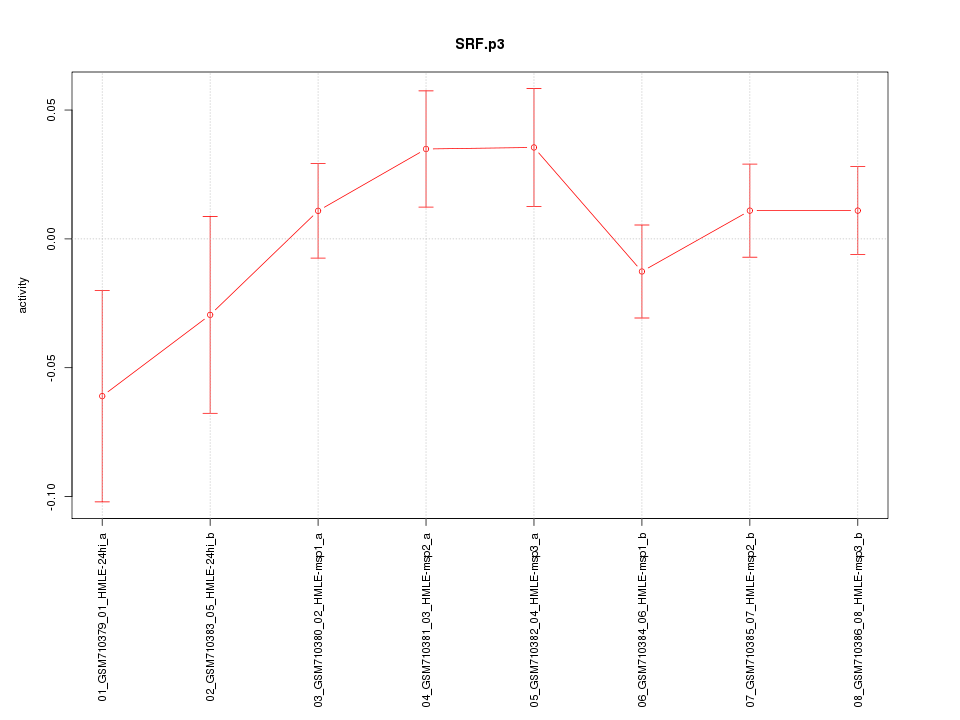
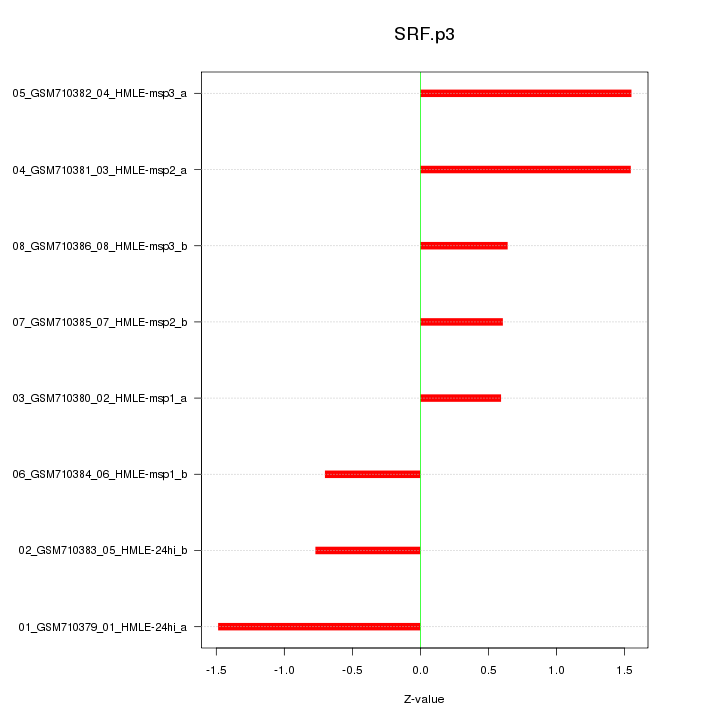
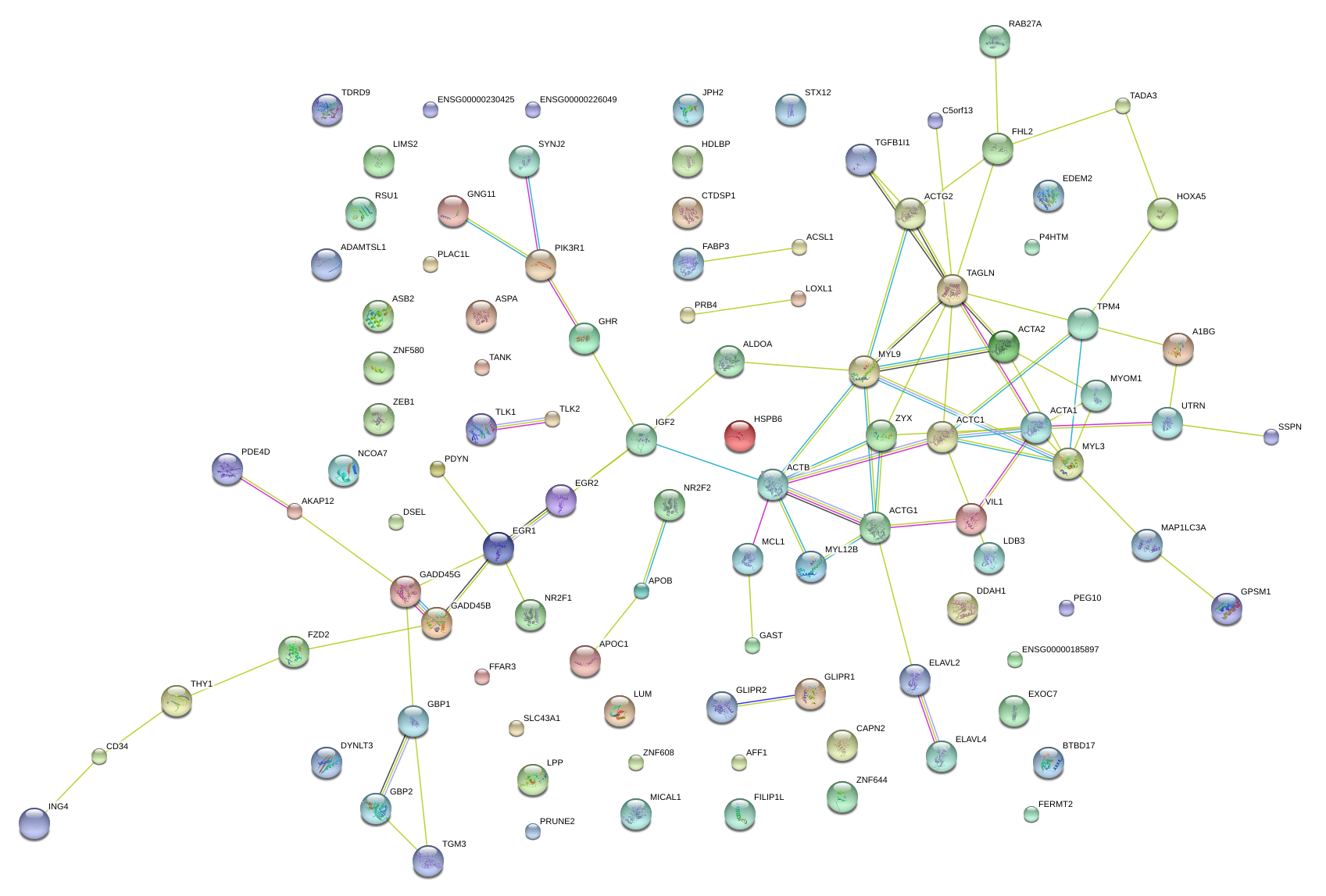
|
chr20_+_35169876
|
3.843
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr20_+_35169897
|
3.229
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr10_-_64576123
|
2.311
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr10_-_64576074
|
2.250
|
|
EGR2
|
early growth response 2
|
|
chr3_-_151987332
|
1.968
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr5_+_137801166
|
1.766
|
|
EGR1
|
early growth response 1
|
|
chr9_+_36136710
|
1.386
|
NM_022343
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr11_+_117070039
|
1.379
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr19_+_2476122
|
1.306
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr5_+_137801178
|
1.256
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr9_-_79520878
|
1.240
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr10_+_31610063
|
1.095
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr14_-_53417808
|
1.012
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr5_+_92918924
|
0.978
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr9_+_92219926
|
0.885
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr10_-_90712510
|
0.634
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr11_-_119294242
|
0.629
|
NM_006288
|
THY1
|
Thy-1 cell surface antigen
|
|
chr20_+_33146500
|
0.610
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr7_-_27183225
|
0.564
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr3_-_46904879
|
0.550
|
NM_000258
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow
|
|
chr17_+_3379295
|
0.534
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr20_-_33735060
|
0.517
|
NM_001145025
NM_018217
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr1_-_89530890
|
0.464
|
NM_002053
|
GBP1
|
guanylate binding protein 1, interferon-inducible
|
|
chr5_-_124080773
|
0.456
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr1_+_223889253
|
0.445
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr12_-_6772252
|
0.410
|
NM_001127582
NM_001127583
NM_001127584
NM_001127585
NM_001127586
NM_016162
|
ING4
|
inhibitor of growth family, member 4
|
|
chr7_+_143078358
|
0.407
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chr11_-_2162167
|
0.405
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr12_+_26348488
|
0.398
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr3_-_99594915
|
0.394
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr11_-_119293848
|
0.389
|
|
THY1
|
Thy-1 cell surface antigen
|
|
chr17_-_72357957
|
0.378
|
NM_001080466
|
BTBD17
|
BTB (POZ) domain containing 17
|
|
chr7_+_143078399
|
0.375
|
|
ZYX
|
zyxin
|
|
chr2_-_128432642
|
0.368
|
NM_001136037
NM_001161403
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr19_-_36247917
|
0.368
|
NM_144617
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chrX_-_37706816
|
0.359
|
NM_006520
|
DYNLT3
|
dynein, light chain, Tctex-type 3
|
|
chr3_+_187930720
|
0.358
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr7_+_143078444
|
0.348
|
|
ZYX
|
zyxin
|
|
chr7_+_143078447
|
0.348
|
|
ZYX
|
zyxin
|
|
chr2_+_219283809
|
0.345
|
NM_007127
|
VIL1
|
villin 1
|
|
chr7_+_143078395
|
0.341
|
|
ZYX
|
zyxin
|
|
chr5_-_58295758
|
0.337
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_28099682
|
0.332
|
NM_177424
|
STX12
|
syntaxin 12
|
|
chr5_-_111093251
|
0.320
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr7_+_93551015
|
0.308
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr5_+_140749830
|
0.304
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr14_+_104394751
|
0.289
|
NM_153046
|
TDRD9
|
tudor domain containing 9
|
|
chr7_+_143078457
|
0.287
|
|
ZYX
|
zyxin
|
|
chr2_-_106015423
|
0.286
|
NM_001039492
NM_001450
NM_201555
|
FHL2
|
four and a half LIM domains 2
|
|
chr2_+_87754991
|
0.285
|
|
LINC00152
|
long intergenic non-protein coding RNA 152
|
|
chr12_+_75874512
|
0.283
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr6_+_151561094
|
0.281
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr1_-_86043932
|
0.273
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_-_172087823
|
0.272
|
NM_001136554
|
TLK1
|
tousled-like kinase 1
|
|
chr2_-_242212244
|
0.264
|
NM_001243900
NM_203346
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr15_-_35087748
|
0.264
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr9_+_139249251
|
0.259
|
NM_001200003
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr6_-_109775423
|
0.257
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr19_+_16178170
|
0.254
|
NM_001145160
|
TPM4
|
tropomyosin 4
|
|
chr19_+_45417576
|
0.242
|
|
APOC1
|
apolipoprotein C-I
|
|
chr5_+_42565562
|
0.240
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr4_-_185747051
|
0.240
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr5_+_42565963
|
0.237
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr1_-_31845818
|
0.235
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr15_+_74218962
|
0.235
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr15_-_55581969
|
0.213
|
NM_183235
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr19_+_56153417
|
0.210
|
NM_016202
NM_001163423
|
ZNF580
|
zinc finger protein 580
|
|
chr18_-_65182507
|
0.200
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr2_-_242212198
|
0.197
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr10_-_16859389
|
0.195
|
|
RSU1
|
Ras suppressor protein 1
|
|
chr6_+_144612872
|
0.193
|
NM_007124
|
UTRN
|
utrophin
|
|
chr1_-_208084432
|
0.187
|
NM_001025109
NM_001773
|
CD34
|
CD34 molecule
|
|
chr11_+_59807747
|
0.181
|
NM_173801
|
PLAC1L
|
placenta-specific 1-like
|
|
chr12_-_11463353
|
0.173
|
NM_002723
|
PRB4
|
proline-rich protein BstNI subfamily 4
|
|
chr12_-_91505541
|
0.173
|
NM_002345
|
LUM
|
lumican
|
|
chr15_+_96869156
|
0.171
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr19_+_35849487
|
0.171
|
NM_005304
|
FFAR3
|
free fatty acid receptor 3
|
|
chr9_+_18474078
|
0.170
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr7_+_143078608
|
0.169
|
|
ZYX
|
zyxin
|
|
chr2_+_74120092
|
0.168
|
NM_001199893
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr3_-_9834357
|
0.164
|
NM_006354
NM_133480
|
TADA3
|
transcriptional adaptor 3
|
|
chr7_+_94285676
|
0.163
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr16_+_30077120
|
0.161
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr1_+_50574601
|
0.159
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr20_+_2276612
|
0.154
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr17_+_39868577
|
0.152
|
NM_000805
|
GAST
|
gastrin
|
|
chr1_-_150552077
|
0.151
|
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr5_+_67588395
|
0.151
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr2_+_161993465
|
0.151
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr2_-_21266915
|
0.149
|
NM_000384
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr3_+_49027281
|
0.149
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr20_-_1974393
|
0.148
|
NM_001190900
|
PDYN
|
prodynorphin
|
|
chr16_+_30077146
|
0.147
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr2_+_74120132
|
0.143
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr1_-_89591685
|
0.141
|
NM_004120
|
GBP2
|
guanylate binding protein 2, interferon-inducible
|
|
chr2_+_87755059
|
0.140
|
|
LINC00152
|
long intergenic non-protein coding RNA 152
|
|
chr6_+_158402928
|
0.139
|
|
SYNJ2
|
synaptojanin 2
|
|
chr11_-_57282315
|
0.137
|
NM_001198810
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chr7_+_94285736
|
0.137
|
|
PEG10
|
paternally expressed 10
|
|
chr2_+_219264477
|
0.136
|
NM_021198
NM_182642
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr18_-_3219846
|
0.135
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr1_-_91487769
|
0.134
|
NM_032186
NM_201269
|
ZNF644
|
zinc finger protein 644
|
|
chr3_+_187871473
|
0.133
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr6_+_126240369
|
0.131
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr10_+_88428167
|
0.128
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr17_-_74099855
|
0.127
|
NM_001013839
NM_001145297
NM_001145298
NM_001145299
NM_015219
|
EXOC7
|
exocyst complex component 7
|
|
chr14_-_94443065
|
0.127
|
NM_001202429
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr16_+_31483066
|
0.123
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr19_-_58858900
|
0.120
|
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr20_-_42815732
|
0.119
|
|
JPH2
|
junctophilin 2
|
|
chr10_-_16859336
|
0.118
|
|
RSU1
|
Ras suppressor protein 1
|
|
chr17_+_42634598
|
0.118
|
NM_001466
|
FZD2
|
frizzled family receptor 2
|
|
chr4_+_87968191
|
0.117
|
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr20_-_33872505
|
0.116
|
NM_181468
|
EIF6
|
eukaryotic translation initiation factor 6
|
|
chr1_+_6105980
|
0.113
|
NM_001199862
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr2_+_207024614
|
0.112
|
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr9_-_98269686
|
0.111
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr19_+_57901330
|
0.111
|
|
ZNF548
ZNF17
|
zinc finger protein 548
zinc finger protein 17
|
|
chr3_-_9834065
|
0.111
|
|
TADA3
|
transcriptional adaptor 3
|
|
chr4_-_10118444
|
0.109
|
|
WDR1
|
WD repeat domain 1
|
|
chrX_-_153602928
|
0.108
|
|
FLNA
|
filamin A, alpha
|
|
chr10_-_16859408
|
0.108
|
NM_012425
NM_152724
|
RSU1
|
Ras suppressor protein 1
|
|
chr1_-_91487593
|
0.106
|
|
ZNF644
|
zinc finger protein 644
|
|
chr19_+_13135368
|
0.105
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr7_+_134464161
|
0.104
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr9_+_97521930
|
0.104
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr9_+_116356199
|
0.104
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr20_-_1974701
|
0.102
|
NM_001190898
NM_001190899
NM_024411
|
PDYN
|
prodynorphin
|
|
chr16_+_30076993
|
0.100
|
NM_001243177
NM_184041
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr20_-_21494663
|
0.095
|
NM_002509
|
NKX2-2
|
NK2 homeobox 2
|
|
chr21_-_47704166
|
0.095
|
|
MCM3AP
|
minichromosome maintenance complex component 3 associated protein
|
|
chr10_+_88428263
|
0.091
|
NM_001080116
NM_001080114
NM_001080115
NM_001171611
NM_007078
|
LDB3
|
LIM domain binding 3
|
|
chr5_-_39425297
|
0.090
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr4_-_10118460
|
0.088
|
|
WDR1
|
WD repeat domain 1
|
|
chr15_+_96873845
|
0.087
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr16_-_67977956
|
0.085
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr12_-_11548485
|
0.084
|
NM_006248
|
PRB1
PRB2
|
proline-rich protein BstNI subfamily 1
proline-rich protein BstNI subfamily 2
|
|
chr3_-_121379757
|
0.083
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr9_-_98269480
|
0.080
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr5_+_176875325
|
0.080
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr17_+_16318911
|
0.080
|
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr11_+_58390145
|
0.079
|
NM_000614
|
CNTF
|
ciliary neurotrophic factor
|
|
chr19_+_42082530
|
0.079
|
NM_001098506
NM_033543
|
CEACAM21
|
carcinoembryonic antigen-related cell adhesion molecule 21
|
|
chr2_-_135476541
|
0.079
|
NM_030923
|
TMEM163
|
transmembrane protein 163
|
|
chr1_+_219347195
|
0.079
|
|
LYPLAL1
|
lysophospholipase-like 1
|
|
chr21_-_28217272
|
0.078
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr4_-_140060601
|
0.078
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr4_-_88450601
|
0.078
|
NM_001128310
NM_004684
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr1_+_219347123
|
0.077
|
NM_138794
|
LYPLAL1
|
lysophospholipase-like 1
|
|
chr19_+_57901188
|
0.076
|
NM_001172773
NM_152909
|
ZNF548
|
zinc finger protein 548
|
|
chr3_-_157221362
|
0.076
|
NM_001167915
NM_001167916
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr8_+_37620100
|
0.076
|
NM_007198
|
PROSC
|
proline synthetase co-transcribed homolog (bacterial)
|
|
chr2_+_219264719
|
0.074
|
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr6_+_158402916
|
0.073
|
|
SYNJ2
|
synaptojanin 2
|
|
chr2_+_85132762
|
0.072
|
NM_021103
|
TMSB10
|
thymosin beta 10
|
|
chr3_+_9834778
|
0.072
|
NM_001198780
|
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa
|
|
chr14_-_106111114
|
0.070
|
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker)
|
|
chr12_+_113860146
|
0.070
|
NM_138432
|
SDSL
|
serine dehydratase-like
|
|
chr16_-_11375094
|
0.069
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr19_+_1026608
|
0.069
|
|
CNN2
|
calponin 2
|
|
chr9_+_116356407
|
0.069
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr12_-_8765438
|
0.069
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr7_+_12726413
|
0.068
|
NM_001037164
NM_212460
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr17_+_16318848
|
0.067
|
NM_016113
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr1_-_935469
|
0.067
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr6_-_86388419
|
0.067
|
|
SNHG5
|
small nucleolar RNA host gene 5 (non-protein coding)
|
|
chr17_-_79479780
|
0.067
|
|
ACTG1
|
actin, gamma 1
|
|
chr19_-_46283763
|
0.065
|
NM_001081563
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr17_-_41132019
|
0.064
|
NM_025267
|
AARSD1
|
alanyl-tRNA synthetase domain containing 1
|
|
chr1_+_244816434
|
0.063
|
|
PPPDE1
|
PPPDE peptidase domain containing 1
|
|
chr22_-_43343046
|
0.062
|
NM_007229
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr17_-_1389013
|
0.061
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr19_+_45417920
|
0.060
|
NM_001645
|
APOC1
|
apolipoprotein C-I
|
|
chr2_+_131513007
|
0.060
|
NM_152698
NM_001105193
|
FAM123C
|
family with sequence similarity 123C
|
|
chr17_-_78450247
|
0.060
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr17_+_80193649
|
0.060
|
NM_001206952
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr17_+_33474835
|
0.059
|
NM_001033576
NM_173167
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chrX_-_153602998
|
0.059
|
NM_001110556
NM_001456
|
FLNA
|
filamin A, alpha
|
|
chr2_-_21266893
|
0.059
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr11_-_19223516
|
0.057
|
NM_001127656
NM_003476
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein)
|
|
chr14_-_74226634
|
0.056
|
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr7_+_114055048
|
0.054
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr3_+_119187782
|
0.053
|
NM_152305
|
POGLUT1
|
protein O-glucosyltransferase 1
|
|
chr3_-_178984789
|
0.051
|
NM_001163677
NM_171828
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr12_+_6881669
|
0.050
|
NM_002286
|
LAG3
|
lymphocyte-activation gene 3
|
|
chr17_+_33914320
|
0.047
|
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr19_-_42894419
|
0.046
|
NM_032488
|
CNFN
|
cornifelin
|
|
chr18_-_47806361
|
0.046
|
NM_001204151
|
MBD1
|
methyl-CpG binding domain protein 1
|
|
chr1_-_89488412
|
0.046
|
NM_018284
|
GBP3
|
guanylate binding protein 3
|
|
chr11_-_85780899
|
0.045
|
NM_001206947
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr17_-_4890664
|
0.045
|
NM_001171167
NM_001171168
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr15_-_102029872
|
0.044
|
|
PCSK6
|
proprotein convertase subtilisin/kexin type 6
|
|
chr2_-_27531113
|
0.043
|
NM_003353
|
UCN
|
urocortin
|
|
chr11_+_34073188
|
0.043
|
NM_005898
NM_203364
|
CAPRIN1
|
cell cycle associated protein 1
|
|
chr1_-_150552005
|
0.043
|
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr1_-_935352
|
0.043
|
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr3_-_71294229
|
0.043
|
NM_001244814
|
FOXP1
|
forkhead box P1
|
|
chr11_+_75273245
|
0.043
|
|
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1)
|
|
chr18_-_37380249
|
0.042
|
|
LOC647946
|
uncharacterized LOC647946
|
|
chr4_-_10118500
|
0.042
|
NM_005112
NM_017491
|
WDR1
|
WD repeat domain 1
|