|
chr1_+_153003677
|
3.980
|
NM_003125
|
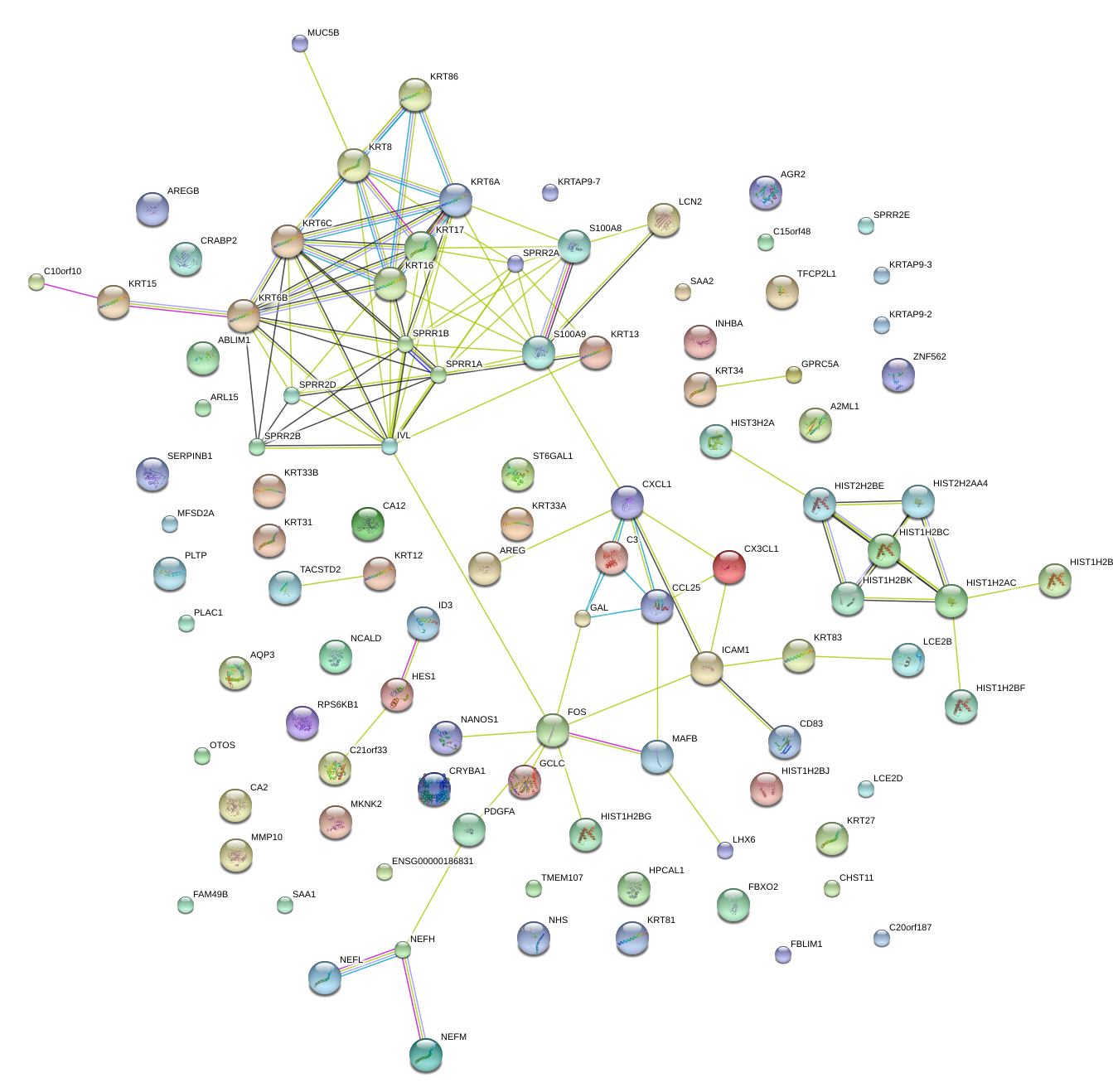
SPRR1B
|
small proline-rich protein 1B
|
|
chr12_-_52886971
|
3.525
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr12_-_52845862
|
3.286
|
NM_005555
|
KRT6B
|
keratin 6B
|
|
chr12_-_52867521
|
2.737
|
NM_173086
|
KRT6C
|
keratin 6C
|
|
chr11_+_18287807
|
2.450
|
NM_000331
NM_001178006
NM_199161
|
SAA1
|
serum amyloid A1
|
|
chr1_-_153363544
|
2.417
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr11_-_18270167
|
2.290
|
NM_001199744
NM_001127380
NM_030754
|
SAA2-SAA4
SAA2
|
SAA2-SAA4 readthrough
serum amyloid A2
|
|
chr9_-_33447527
|
2.278
|
NM_004925
|
AQP3
|
aquaporin 3 (Gill blood group)
|
|
chr19_-_6720585
|
2.208
|
NM_000064
|
C3
|
complement component 3
|
|
chr1_+_153330329
|
2.162
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr1_-_153029987
|
2.058
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr17_-_39507055
|
1.858
|
NM_004138
|
KRT33A
|
keratin 33A
|
|
chr11_-_102651321
|
1.823
|
NM_002425
|
MMP10
|
matrix metallopeptidase 10 (stromelysin 2)
|
|
chr14_+_75745480
|
1.775
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr3_+_193853930
|
1.726
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr17_-_39780762
|
1.646
|
NM_000422
|
KRT17
|
keratin 17
|
|
chr15_-_63674074
|
1.561
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chr1_+_152881022
|
1.555
|
NM_005547
|
IVL
|
involucrin
|
|
chr1_-_59042827
|
1.548
|
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr15_-_63673918
|
1.516
|
|
CA12
|
carbonic anhydrase XII
|
|
chr3_+_193853937
|
1.452
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr8_+_24771276
|
1.427
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr12_-_52685249
|
1.388
|
NM_002281
|
KRT81
|
keratin 81
|
|
chr8_+_24771268
|
1.385
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr6_-_2842069
|
1.384
|
NM_030666
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr12_+_13044352
|
1.381
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr8_+_86376130
|
1.353
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr22_+_29876180
|
1.316
|
NM_021076
|
NEFH
|
neurofilament, heavy polypeptide
|
|
chr2_-_122042767
|
1.310
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr1_-_156675237
|
1.202
|
NM_001878
NM_001199723
|
CRABP2
|
cellular retinoic acid binding protein 2
|
|
chr6_+_26124372
|
1.176
|
NM_003512
|
HIST1H2AC
|
histone cluster 1, H2ac
|
|
chr12_-_53298765
|
1.174
|
NM_002273
|
KRT8
|
keratin 8
|
|
chr1_-_153013593
|
1.158
|
NM_006945
|
SPRR2D
|
small proline-rich protein 2D
|
|
chr17_-_38938785
|
1.140
|
NM_181537
|
KRT27
|
keratin 27
|
|
chr4_+_75310852
|
1.134
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr10_-_45474210
|
1.112
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr17_-_39675078
|
1.112
|
NM_002275
|
KRT15
|
keratin 15
|
|
chr17_+_39382878
|
1.109
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr10_-_116418056
|
1.094
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr8_-_24814028
|
1.071
|
NM_006158
|
NEFL
|
neurofilament, light polypeptide
|
|
chr1_-_11714471
|
1.038
|
|
FBXO2
|
F-box protein 2
|
|
chr1_-_23885916
|
1.006
|
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein
|
|
chrX_-_133792497
|
1.003
|
NM_021796
|
PLAC1
|
placenta-specific 1
|
|
chr7_-_41742666
|
1.001
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr17_-_39768939
|
0.992
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr20_-_44540685
|
0.988
|
|
PLTP
|
phospholipid transfer protein
|
|
chr1_-_153067000
|
0.978
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr12_+_8975149
|
0.904
|
NM_144670
|
A2ML1
|
alpha-2-macroglobulin-like 1
|
|
chr19_+_10381763
|
0.882
|
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr1_+_40420802
|
0.876
|
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr6_-_26216871
|
0.869
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr10_+_120789227
|
0.769
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr3_+_186739643
|
0.763
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chrX_+_17393537
|
0.735
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr9_-_124989755
|
0.728
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr1_-_11714396
|
0.727
|
|
FBXO2
|
F-box protein 2
|
|
chr6_-_53409897
|
0.726
|
NM_001197115
NM_001498
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr19_+_8117645
|
0.724
|
NM_001201359
NM_005624
|
CCL25
|
chemokine (C-C motif) ligand 25
|
|
chr17_-_39661803
|
0.714
|
NM_002274
NM_153490
|
KRT13
|
keratin 13
|
|
chr6_-_27114577
|
0.714
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr1_-_153044083
|
0.711
|
NM_001017418
|
SPRR2B
|
small proline-rich protein 2B
|
|
chr9_+_130911731
|
0.690
|
NM_005564
|
LCN2
|
lipocalin 2
|
|
chr15_+_45722762
|
0.675
|
NM_032413
NM_197955
|
C15orf48
|
chromosome 15 open reading frame 48
|
|
chr16_+_57406398
|
0.672
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr12_+_104850784
|
0.671
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr5_-_53606387
|
0.638
|
NM_019087
|
ARL15
|
ADP-ribosylation factor-like 15
|
|
chr2_-_241080072
|
0.627
|
NM_148961
|
OTOS
|
otospiralin
|
|
chr4_+_74735107
|
0.626
|
NM_001511
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha)
|
|
chr7_-_559479
|
0.623
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr11_+_1151578
|
0.620
|
|
MUC5AC
|
mucin 5AC, oligomeric mucus/gel-forming
|
|
chr19_-_2051222
|
0.602
|
NM_017572
NM_199054
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr8_-_131028674
|
0.593
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr20_-_39317875
|
0.569
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr1_+_152658598
|
0.559
|
NM_014357
|
LCE2B
|
late cornified envelope 2B
|
|
chr17_+_27573874
|
0.549
|
NM_005208
|
CRYBA1
|
crystallin, beta A1
|
|
chr1_+_16085245
|
0.532
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr7_-_16844606
|
0.531
|
NM_006408
|
AGR2
|
anterior gradient 2 homolog (Xenopus laevis)
|
|
chr6_+_14118002
|
0.529
|
|
CD83
|
CD83 molecule
|
|
chr11_+_68452007
|
0.527
|
|
GAL
|
galanin prepropeptide
|
|
chr17_-_39023461
|
0.519
|
NM_000223
|
KRT12
|
keratin 12
|
|
chr16_+_78056411
|
0.513
|
NM_005752
|
CLEC3A
|
C-type lectin domain family 3, member A
|
|
chr14_-_55369163
|
0.510
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr8_-_131028646
|
0.499
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr10_+_24497719
|
0.494
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr15_-_75017710
|
0.494
|
NM_000499
|
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1
|
|
chr19_+_12902295
|
0.490
|
NM_002229
|
JUNB
|
jun B proto-oncogene
|
|
chr19_-_51487070
|
0.487
|
NM_139277
NM_001207053
NM_001243126
NM_005046
|
KLK7
|
kallikrein-related peptidase 7
|
|
chr3_+_70048882
|
0.486
|
|
|
|
|
chr12_+_101988746
|
0.481
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr17_-_48546174
|
0.481
|
NM_001267
|
CHAD
|
chondroadherin
|
|
chr12_+_13044493
|
0.480
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr6_-_26124130
|
0.472
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr7_-_25268057
|
0.470
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr9_-_37034358
|
0.464
|
NM_016734
|
PAX5
|
paired box 5
|
|
chr17_+_56315786
|
0.464
|
NM_001160102
NM_006151
|
LPO
|
lactoperoxidase
|
|
chr19_+_13049428
|
0.460
|
|
CALR
|
calreticulin
|
|
chr19_-_33793441
|
0.442
|
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr1_+_42619091
|
0.438
|
NM_007102
|
GUCA2B
|
guanylate cyclase activator 2B (uroguanylin)
|
|
chr11_-_5248180
|
0.436
|
NM_000518
|
HBB
|
hemoglobin, beta
|
|
chr1_-_23886284
|
0.433
|
NM_002167
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein
|
|
chr1_+_37940180
|
0.426
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr4_-_46391393
|
0.425
|
NM_001114175
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2
|
|
chr6_-_133035180
|
0.422
|
NM_004666
|
VNN1
|
vanin 1
|
|
chr19_+_13049413
|
0.417
|
NM_004343
|
CALR
|
calreticulin
|
|
chr17_+_32612686
|
0.416
|
NM_002986
|
CCL11
|
chemokine (C-C motif) ligand 11
|
|
chr8_-_48650703
|
0.415
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr1_-_228135587
|
0.415
|
NM_003395
|
WNT9A
|
wingless-type MMTV integration site family, member 9A
|
|
chr1_-_27481400
|
0.413
|
NM_003047
|
SLC9A1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1
|
|
chrX_+_84498993
|
0.409
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr19_+_8117887
|
0.408
|
|
CCL25
|
chemokine (C-C motif) ligand 25
|
|
chr22_-_27013959
|
0.396
|
NM_001887
|
CRYBB1
|
crystallin, beta B1
|
|
chr14_-_103989195
|
0.393
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chr19_+_13049440
|
0.383
|
|
CALR
|
calreticulin
|
|
chr19_-_35981345
|
0.383
|
NM_001244847
NM_207392
|
KRTDAP
|
keratinocyte differentiation-associated protein
|
|
chr6_-_27100528
|
0.381
|
NM_021058
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr3_-_27764197
|
0.367
|
|
EOMES
|
eomesodermin
|
|
chr12_+_41086189
|
0.363
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr1_-_43424536
|
0.357
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr6_+_127898318
|
0.355
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr21_-_32185560
|
0.353
|
NM_175857
|
KRTAP8-1
|
keratin associated protein 8-1
|
|
chr17_+_7387688
|
0.347
|
NM_000937
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa
|
|
chr19_+_859658
|
0.346
|
NM_001928
|
CFD
|
complement factor D (adipsin)
|
|
chr17_-_39684499
|
0.344
|
|
KRT19
|
keratin 19
|
|
chr19_+_41222915
|
0.344
|
NM_025194
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|
|
chr6_-_2903461
|
0.342
|
NM_004155
|
SERPINB9
|
serpin peptidase inhibitor, clade B (ovalbumin), member 9
|
|
chr11_+_1860232
|
0.342
|
NM_003282
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr4_+_119199906
|
0.332
|
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding)
|
|
chr19_-_11591439
|
0.332
|
NM_001420
NM_032281
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr14_+_94492747
|
0.330
|
|
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2
|
|
chr15_-_62457481
|
0.330
|
NM_001007595
|
C2CD4B
|
C2 calcium-dependent domain containing 4B
|
|
chr10_+_118380451
|
0.330
|
NM_005396
|
PNLIPRP2
|
pancreatic lipase-related protein 2
|
|
chr19_-_33793429
|
0.330
|
NM_004364
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr10_-_69455926
|
0.328
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr15_-_85201417
|
0.319
|
|
NMB
|
neuromedin B
|
|
chr6_+_33172418
|
0.317
|
NM_014234
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8
|
|
chr11_+_68451978
|
0.313
|
NM_015973
|
GAL
|
galanin prepropeptide
|
|
chr1_+_149822627
|
0.312
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr7_+_116593354
|
0.311
|
NM_018412
NM_021908
|
ST7
|
suppression of tumorigenicity 7
|
|
chr4_+_74301932
|
0.310
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr5_+_132209354
|
0.309
|
NM_052971
|
LEAP2
|
liver expressed antimicrobial peptide 2
|
|
chr8_-_48650683
|
0.307
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr1_+_17575592
|
0.306
|
NM_016233
|
PADI3
|
peptidyl arginine deiminase, type III
|
|
chr16_-_3030473
|
0.298
|
NM_004203
NM_182687
|
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1
|
|
chr21_+_34398210
|
0.295
|
NM_005806
|
OLIG2
|
oligodendrocyte lineage transcription factor 2
|
|
chr8_-_23261626
|
0.294
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr1_-_32801606
|
0.292
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr12_+_104850750
|
0.287
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr16_+_71392615
|
0.284
|
NM_001740
NM_007088
|
CALB2
|
calbindin 2
|
|
chr1_-_149858114
|
0.282
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr3_+_148583042
|
0.280
|
NM_001870
|
CPA3
|
carboxypeptidase A3 (mast cell)
|
|
chr17_+_34431219
|
0.280
|
NM_002984
|
CCL4
|
chemokine (C-C motif) ligand 4
|
|
chr6_-_26197477
|
0.279
|
|
HIST1H3D
|
histone cluster 1, H3d
|
|
chr7_+_115863004
|
0.277
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr8_+_133879202
|
0.276
|
NM_003235
|
TG
|
thyroglobulin
|
|
chr1_+_6105980
|
0.275
|
NM_001199862
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr14_-_103989112
|
0.273
|
|
CKB
|
creatine kinase, brain
|
|
chr2_+_113885137
|
0.272
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr8_-_23261591
|
0.267
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr20_+_2276612
|
0.266
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr16_+_71392637
|
0.263
|
|
CALB2
|
calbindin 2
|
|
chr17_+_79008945
|
0.263
|
NM_001144888
NM_006340
NM_017450
NM_017451
|
BAIAP2
|
BAI1-associated protein 2
|
|
chr17_+_7554239
|
0.261
|
NM_001678
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr6_-_27775683
|
0.260
|
NM_003519
|
HIST1H2BL
|
histone cluster 1, H2bl
|
|
chr17_-_39580657
|
0.259
|
NM_003770
|
KRT37
|
keratin 37
|
|
chr17_+_39394269
|
0.259
|
NM_031963
|
KRTAP9-8
|
keratin associated protein 9-8
|
|
chr6_+_111195986
|
0.257
|
NM_001634
NM_001033059
|
AMD1
|
adenosylmethionine decarboxylase 1
|
|
chr11_-_57148602
|
0.257
|
NM_006093
|
PRG3
|
proteoglycan 3
|
|
chr11_-_5255712
|
0.256
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr11_+_65190272
|
0.255
|
|
|
|
|
chr11_+_66188474
|
0.252
|
NM_178864
|
NPAS4
|
neuronal PAS domain protein 4
|
|
chr2_-_75426613
|
0.251
|
NM_001058
NM_015727
|
TACR1
|
tachykinin receptor 1
|
|
chr1_-_149783910
|
0.250
|
NM_001024599
NM_001161334
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr1_-_149814264
|
0.247
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr1_+_233749749
|
0.246
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr5_-_57755787
|
0.245
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chrX_+_153046422
|
0.244
|
NM_001170760
NM_001170761
NM_014370
|
SRPK3
|
SRSF protein kinase 3
|
|
chr7_-_16505292
|
0.242
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chrX_-_30327479
|
0.241
|
NM_000475
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr3_+_4535030
|
0.240
|
NM_001099952
NM_001168272
NM_002222
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1
|
|
chr1_-_8086347
|
0.237
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr8_+_1993157
|
0.234
|
NM_003970
|
MYOM2
|
myomesin (M-protein) 2, 165kDa
|
|
chr1_-_203155813
|
0.232
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr15_+_45406518
|
0.226
|
NM_207581
|
DUOXA2
|
dual oxidase maturation factor 2
|
|
chr2_-_142889269
|
0.225
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr8_-_23261632
|
0.222
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr10_-_95170
|
0.222
|
NM_177987
|
TUBB8
|
tubulin, beta 8 class VIII
|
|
chr18_-_44556448
|
0.222
|
NM_145653
|
TCEB3C
|
transcription elongation factor B polypeptide 3C (elongin A3)
|
|
chr9_-_117568292
|
0.221
|
NM_005118
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr9_+_139557378
|
0.217
|
NM_016215
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr7_+_100612903
|
0.215
|
NM_001164462
|
MUC12
|
mucin 12, cell surface associated
|
|
chr1_+_37940129
|
0.213
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr12_+_52400725
|
0.212
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr1_+_247581350
|
0.212
|
NM_001127462
NM_001243133
NM_004895
NM_183395
|
NLRP3
|
NLR family, pyrin domain containing 3
|
|
chr17_+_79008958
|
0.208
|
|
BAIAP2
|
BAI1-associated protein 2
|
|
chr1_-_26393984
|
0.207
|
NM_032588
|
TRIM63
|
tripartite motif containing 63
|
|
chr7_+_116593554
|
0.206
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr16_+_56966025
|
0.203
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr6_-_137815520
|
0.203
|
NM_175747
|
OLIG3
|
oligodendrocyte transcription factor 3
|
|
chr1_-_203155827
|
0.203
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr7_+_94285676
|
0.202
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|