|
chr7_+_94023872
|
7.158
|
NM_000089
|
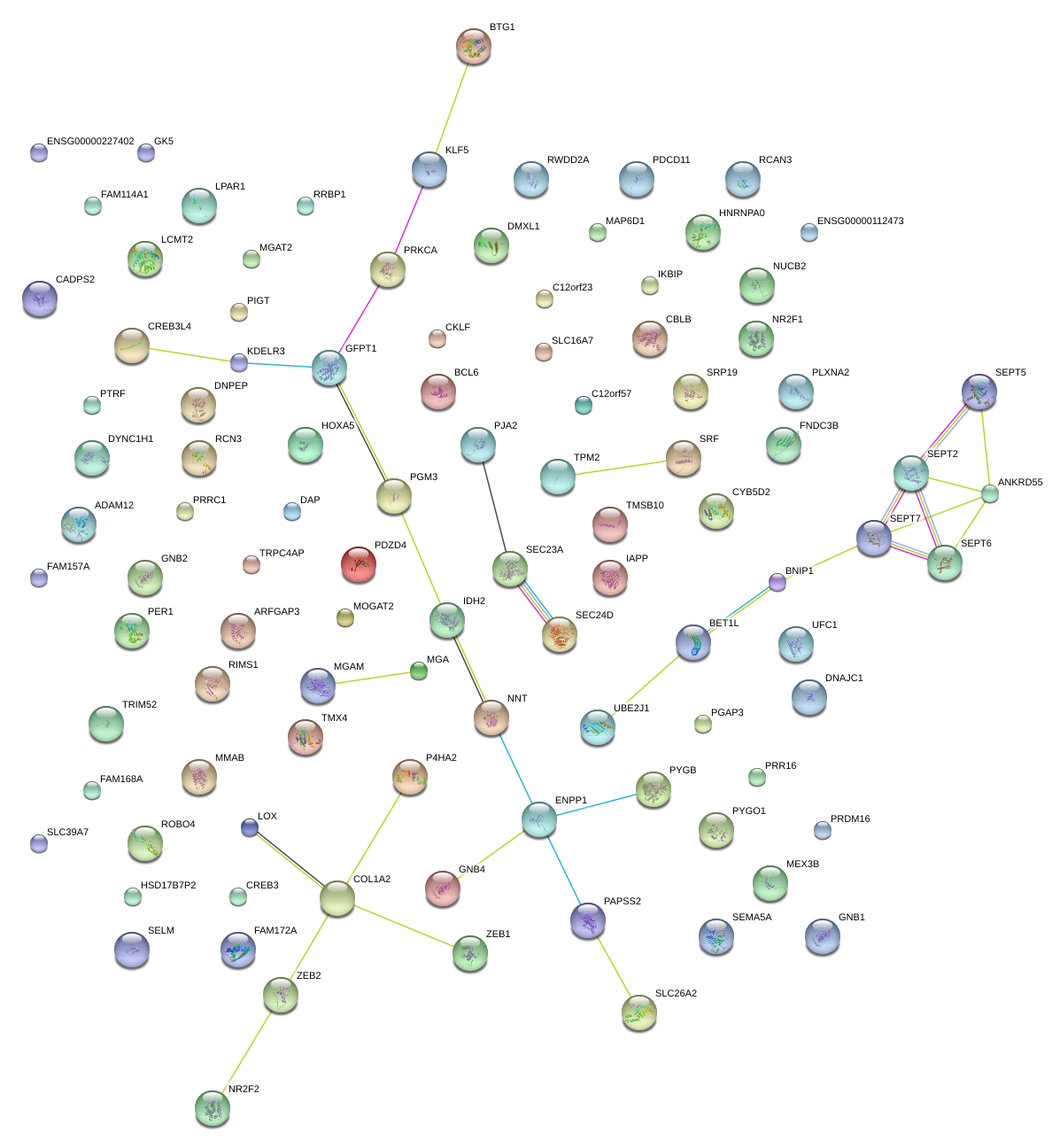
COL1A2
|
collagen, type I, alpha 2
|
|
chr19_+_50031232
|
4.059
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr5_+_119800004
|
3.200
|
NM_016644
|
PRR16
|
proline rich 16
|
|
chr5_-_9546157
|
3.133
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr7_+_94024201
|
3.113
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr5_-_121413906
|
2.761
|
|
LOX
|
lysyl oxidase
|
|
chr22_+_38864015
|
2.697
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr5_+_119799898
|
2.694
|
|
PRR16
|
proline rich 16
|
|
chr5_-_121414011
|
2.243
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr6_+_72892371
|
2.224
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr10_+_89419352
|
2.076
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr19_+_50030874
|
1.742
|
NM_020650
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr5_+_92920592
|
1.680
|
|
NR2F1
NR2F2
|
nuclear receptor subfamily 2, group F, member 1
nuclear receptor subfamily 2, group F, member 2
|
|
chr15_-_82338459
|
1.491
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr15_-_82338348
|
1.255
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr5_-_121413967
|
1.208
|
|
LOX
|
lysyl oxidase
|
|
chr5_-_10761341
|
1.203
|
|
DAP
|
death-associated protein
|
|
chr5_-_10761344
|
1.135
|
NM_004394
|
DAP
|
death-associated protein
|
|
chr12_-_99038401
|
1.131
|
NM_153687
NM_201612
NM_201613
|
IKBIP
|
IKBKB interacting protein
|
|
chr22_-_31503484
|
1.129
|
NM_080430
|
SELM
|
selenoprotein M
|
|
chr6_+_132129195
|
1.024
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr6_+_83903031
|
1.023
|
NM_033411
|
RWDD2A
|
RWD domain containing 2A
|
|
chr4_+_38869346
|
1.009
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr11_-_2018703
|
1.009
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr15_+_41952641
|
0.956
|
|
MGA
|
MAX gene associated
|
|
chr5_-_10761179
|
0.956
|
|
DAP
|
death-associated protein
|
|
chr3_-_183543295
|
0.944
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr20_+_44044706
|
0.864
|
NM_001184728
NM_001184729
NM_001184730
NM_015937
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis, class T
|
|
chr12_+_7053180
|
0.848
|
NM_138425
|
C12orf57
|
chromosome 12 open reading frame 57
|
|
chr5_+_126853308
|
0.836
|
NM_130809
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr6_+_33168602
|
0.815
|
NM_001077516
NM_006979
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr17_+_4046941
|
0.759
|
|
CYB5D2
|
cytochrome b5 domain containing 2
|
|
chr17_+_64298844
|
0.733
|
NM_002737
|
PRKCA
|
protein kinase C, alpha
|
|
chr5_-_131563481
|
0.723
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr10_-_22292362
|
0.675
|
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr12_+_107349673
|
0.668
|
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr15_-_55880508
|
0.666
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr5_-_137089754
|
0.658
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr12_-_99038705
|
0.657
|
|
IKBIP
|
IKBKB interacting protein
|
|
chr17_-_8059637
|
0.653
|
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr20_+_25228698
|
0.640
|
NM_002862
|
PYGB
|
phosphorylase, glycogen; brain
|
|
chr10_-_128077072
|
0.635
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr6_+_132129154
|
0.626
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr3_+_171758289
|
0.620
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr14_-_39572278
|
0.592
|
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr1_+_161123755
|
0.591
|
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1
|
|
chr5_-_93447237
|
0.586
|
NM_001163417
NM_001163418
NM_032042
|
FAM172A
|
family with sequence similarity 172, member A
|
|
chr5_-_137089434
|
0.579
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr9_+_35732207
|
0.575
|
NM_006368
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr15_-_90645621
|
0.571
|
|
IDH2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial
|
|
chr12_-_92539614
|
0.565
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr9_+_35732498
|
0.553
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr3_-_105587710
|
0.552
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr3_-_179169180
|
0.545
|
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr6_-_83902921
|
0.542
|
NM_001199918
NM_001199919
NM_015599
|
PGM3
|
phosphoglucomutase 3
|
|
chr9_+_35732629
|
0.529
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr22_-_43253186
|
0.527
|
|
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3
|
|
chr11_-_207324
|
0.527
|
|
BET1L
|
blocked early in transport 1 homolog (S. cerevisiae)-like
|
|
chr14_+_50087400
|
0.519
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr20_-_17662704
|
0.508
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr2_-_69614321
|
0.504
|
NM_001244710
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr4_-_119757183
|
0.492
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr9_-_113800243
|
0.491
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr7_-_27183225
|
0.490
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr5_+_112196884
|
0.486
|
NM_001204193
NM_001204194
NM_001204196
NM_001204199
NM_003135
|
SRP19
|
signal recognition particle 19kDa
|
|
chrX_-_118827322
|
0.480
|
NM_015129
NM_145799
NM_145800
NM_145802
|
SEPT6
|
septin 6
|
|
chr10_-_22292640
|
0.463
|
NM_022365
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr12_+_59989847
|
0.456
|
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr5_-_108745569
|
0.453
|
NM_014819
|
PJA2
|
praja ring finger 2
|
|
chr5_+_149340287
|
0.452
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr3_-_141944441
|
0.452
|
NM_001039547
|
GK5
|
glycerol kinase 5 (putative)
|
|
chr4_-_119757288
|
0.447
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr16_+_66586478
|
0.438
|
|
CKLF
|
chemokine-like factor
|
|
chr22_-_43253304
|
0.438
|
NM_001142293
NM_014570
|
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3
|
|
chr17_-_40575117
|
0.436
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr20_-_17662828
|
0.433
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr2_+_242254722
|
0.432
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr6_-_90062610
|
0.428
|
NM_016021
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr7_-_122526410
|
0.427
|
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chr14_+_102430767
|
0.424
|
NM_001376
|
DYNC1H1
|
dynein, cytoplasmic 1, heavy chain 1
|
|
chr10_-_128077011
|
0.417
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr18_-_34409084
|
0.417
|
NM_015476
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr15_-_43622687
|
0.414
|
NM_014793
|
LCMT2
|
leucine carboxyl methyltransferase 2
|
|
chr2_-_145277568
|
0.413
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr5_-_180688095
|
0.412
|
NM_032765
|
TRIM52
|
tripartite motif containing 52
|
|
chr5_+_172571444
|
0.408
|
NM_001205
NM_013978
NM_013979
NM_013980
|
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1
|
|
chr18_-_34408948
|
0.405
|
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr18_-_34408845
|
0.404
|
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr3_-_187463227
|
0.403
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr20_-_33680601
|
0.401
|
NM_015638
NM_199368
|
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr3_-_141944296
|
0.399
|
|
GK5
|
glycerol kinase 5 (putative)
|
|
chr5_+_112196976
|
0.397
|
|
SRP19
|
signal recognition particle 19kDa
|
|
chr14_-_39572414
|
0.393
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr11_-_73309090
|
0.391
|
NM_015159
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr20_-_8000370
|
0.378
|
NM_021156
|
TMX4
|
thioredoxin-related transmembrane protein 4
|
|
chr1_-_208417635
|
0.374
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr5_+_43603287
|
0.374
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr5_-_137088952
|
0.372
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr2_+_85132999
|
0.370
|
|
TMSB10
|
thymosin beta 10
|
|
chr15_-_90645704
|
0.355
|
NM_002168
|
IDH2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial
|
|
chr9_-_35689902
|
0.355
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr11_+_17298278
|
0.354
|
NM_005013
|
NUCB2
|
nucleobindin 2
|
|
chr5_+_118406721
|
0.354
|
|
DMXL1
|
Dmx-like 1
|
|
chr16_+_66586479
|
0.352
|
|
CKLF
|
chemokine-like factor
|
|
chr7_-_19157262
|
0.345
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr1_-_145039631
|
0.345
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr22_-_20850075
|
0.344
|
NM_032775
|
KLHL22
|
kelch-like 22 (Drosophila)
|
|
chr20_-_33680593
|
0.343
|
|
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr19_+_49468587
|
0.341
|
|
FTL
|
ferritin, light polypeptide
|
|
chr19_-_18391440
|
0.340
|
|
|
|
|
chr2_-_15701414
|
0.336
|
|
NBAS
|
neuroblastoma amplified sequence
|
|
chr14_+_102430897
|
0.336
|
|
DYNC1H1
|
dynein, cytoplasmic 1, heavy chain 1
|
|
chr2_-_242254984
|
0.335
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr16_+_66586447
|
0.335
|
NM_001202509
NM_001204098
NM_001204099
NM_001040138
NM_016326
NM_016951
NM_181640
NM_181641
|
CKLF-CMTM1
CKLF
|
CKLF-CMTM1 readthrough
chemokine-like factor
|
|
chr15_-_60690115
|
0.333
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr3_-_73673978
|
0.330
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr11_-_62599518
|
0.329
|
NM_001244666
NM_003164
|
STX5
|
syntaxin 5
|
|
chr12_+_99038918
|
0.328
|
NM_001160
NM_013229
NM_181861
NM_181868
NM_181869
|
APAF1
|
apoptotic peptidase activating factor 1
|
|
chr20_-_33680352
|
0.328
|
|
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr16_-_30546145
|
0.326
|
NM_023931
|
ZNF747
|
zinc finger protein 747
|
|
chr14_+_100259445
|
0.324
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr6_+_42928464
|
0.324
|
NM_018960
|
GNMT
|
glycine N-methyltransferase
|
|
chr5_-_89825292
|
0.324
|
NM_198273
|
LYSMD3
|
LysM, putative peptidoglycan-binding, domain containing 3
|
|
chr11_-_2019070
|
0.323
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr11_-_66056455
|
0.323
|
|
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae)
|
|
chr1_+_6845381
|
0.320
|
NM_001195563
NM_001242701
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr15_+_76135621
|
0.317
|
NM_173469
|
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2
|
|
chr12_-_101801499
|
0.316
|
NM_001177
|
ARL1
|
ADP-ribosylation factor-like 1
|
|
chr15_+_55700722
|
0.315
|
NM_001198784
|
FLJ27352
|
uncharacterized LOC145788
|
|
chr11_+_61560108
|
0.314
|
NM_004111
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr3_-_197686837
|
0.313
|
NM_032263
|
IQCG
|
IQ motif containing G
|
|
chr16_-_30569505
|
0.310
|
NM_001172679
NM_033410
|
ZNF764
|
zinc finger protein 764
|
|
chr11_-_62599447
|
0.307
|
|
STX5
|
syntaxin 5
|
|
chr12_+_69202796
|
0.303
|
|
MDM2
|
Mdm2 p53 binding protein homolog (mouse)
|
|
chr10_-_18948166
|
0.301
|
|
|
|
|
chr6_-_90062349
|
0.297
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr11_-_61560052
|
0.296
|
NM_014206
|
C11orf10
|
chromosome 11 open reading frame 10
|
|
chr5_-_115177261
|
0.295
|
NM_004707
|
ATG12
|
ATG12 autophagy related 12 homolog (S. cerevisiae)
|
|
chr5_-_176730659
|
0.294
|
NM_001031677
NM_130781
|
RAB24
|
RAB24, member RAS oncogene family
|
|
chr2_-_242212244
|
0.293
|
NM_001243900
NM_203346
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr5_+_115177616
|
0.292
|
NM_001284
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit
|
|
chr11_+_61595633
|
0.285
|
NM_004265
|
FADS2
|
fatty acid desaturase 2
|
|
chr18_-_44702735
|
0.282
|
NM_016097
|
IER3IP1
|
immediate early response 3 interacting protein 1
|
|
chr9_-_119449374
|
0.282
|
NM_198186
NM_001184734
NM_001184735
NM_198187
NM_198188
|
ASTN2
|
astrotactin 2
|
|
chr1_-_144931967
|
0.280
|
NM_001002811
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr19_+_49468557
|
0.278
|
NM_000146
|
FTL
|
ferritin, light polypeptide
|
|
chr11_-_66056607
|
0.278
|
NM_020470
|
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae)
|
|
chr12_+_6937764
|
0.276
|
|
LEPREL2
|
leprecan-like 2
|
|
chr20_-_8000254
|
0.275
|
|
TMX4
|
thioredoxin-related transmembrane protein 4
|
|
chr15_-_90645709
|
0.275
|
|
IDH2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial
|
|
chrX_+_102862833
|
0.275
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr1_+_109633373
|
0.274
|
NM_020141
|
TMEM167B
|
transmembrane protein 167B
|
|
chr1_+_145576011
|
0.274
|
|
PIAS3
|
protein inhibitor of activated STAT, 3
|
|
chr20_+_31407696
|
0.274
|
NM_012325
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr17_+_45000483
|
0.273
|
NM_001012511
NM_004287
NM_054022
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr19_-_19030156
|
0.269
|
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chr11_+_62341542
|
0.268
|
|
|
|
|
chr15_-_55700456
|
0.267
|
NM_004748
NM_001204450
NM_001204451
NM_020739
|
CCPG1
|
cell cycle progression 1
|
|
chr6_-_109703553
|
0.266
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr4_-_5891917
|
0.264
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr11_+_69455938
|
0.263
|
|
CCND1
|
cyclin D1
|
|
chr11_-_2018334
|
0.262
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr5_-_114938060
|
0.262
|
NM_021649
|
TICAM2
|
toll-like receptor adaptor molecule 2
|
|
chr17_+_45000493
|
0.261
|
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr6_-_90062439
|
0.257
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr5_-_157285977
|
0.256
|
NM_001195555
NM_001195556
NM_014666
|
CLINT1
|
clathrin interactor 1
|
|
chr7_-_23053674
|
0.255
|
|
FAM126A
|
family with sequence similarity 126, member A
|
|
chr2_-_15701445
|
0.255
|
NM_015909
|
NBAS
|
neuroblastoma amplified sequence
|
|
chr3_+_127771342
|
0.253
|
|
|
|
|
chr2_-_224702200
|
0.252
|
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit
|
|
chr9_-_133454818
|
0.250
|
|
LOC100272217
|
uncharacterized LOC100272217
|
|
chr6_+_28249313
|
0.249
|
NM_001184743
NM_032507
|
PGBD1
|
piggyBac transposable element derived 1
|
|
chr20_+_31407777
|
0.248
|
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr1_+_145575987
|
0.247
|
NM_006099
|
PIAS3
|
protein inhibitor of activated STAT, 3
|
|
chr2_+_85132762
|
0.246
|
NM_021103
|
TMSB10
|
thymosin beta 10
|
|
chr19_+_49468656
|
0.246
|
|
FTL
|
ferritin, light polypeptide
|
|
chr1_-_22109450
|
0.245
|
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr19_+_17326196
|
0.244
|
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae)
|
|
chr15_+_41952572
|
0.244
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr11_+_61560401
|
0.243
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr5_-_121412805
|
0.240
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr6_-_134496833
|
0.234
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr14_-_50087326
|
0.234
|
NM_001001
|
RPL36AL
|
ribosomal protein L36a-like
|
|
chr7_+_128379400
|
0.233
|
|
CALU
|
calumenin
|
|
chr5_-_171881421
|
0.233
|
NM_001017995
|
SH3PXD2B
|
SH3 and PX domains 2B
|
|
chr12_+_14927305
|
0.233
|
|
H2AFJ
|
H2A histone family, member J
|
|
chr7_+_128379428
|
0.231
|
|
CALU
|
calumenin
|
|
chr7_+_128379452
|
0.230
|
|
CALU
|
calumenin
|
|
chr10_+_83635069
|
0.229
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr18_-_44702647
|
0.229
|
|
IER3IP1
|
immediate early response 3 interacting protein 1
|
|
chr20_-_33735060
|
0.224
|
NM_001145025
NM_018217
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr5_+_43603216
|
0.223
|
NM_182977
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr12_-_133405197
|
0.220
|
NM_005895
|
GOLGA3
|
golgin A3
|
|
chr11_+_65292576
|
0.219
|
|
SCYL1
|
SCY1-like 1 (S. cerevisiae)
|
|
chrX_-_153059832
|
0.217
|
NM_004135
NM_174869
|
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma
|
|
chr18_-_44702732
|
0.215
|
|
IER3IP1
|
immediate early response 3 interacting protein 1
|
|
chr12_+_107349539
|
0.215
|
NM_152261
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr19_-_19030195
|
0.214
|
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chr19_-_19030196
|
0.213
|
|
COPE
|
coatomer protein complex, subunit epsilon
|
|
chr13_-_36920777
|
0.213
|
NM_001142295
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|