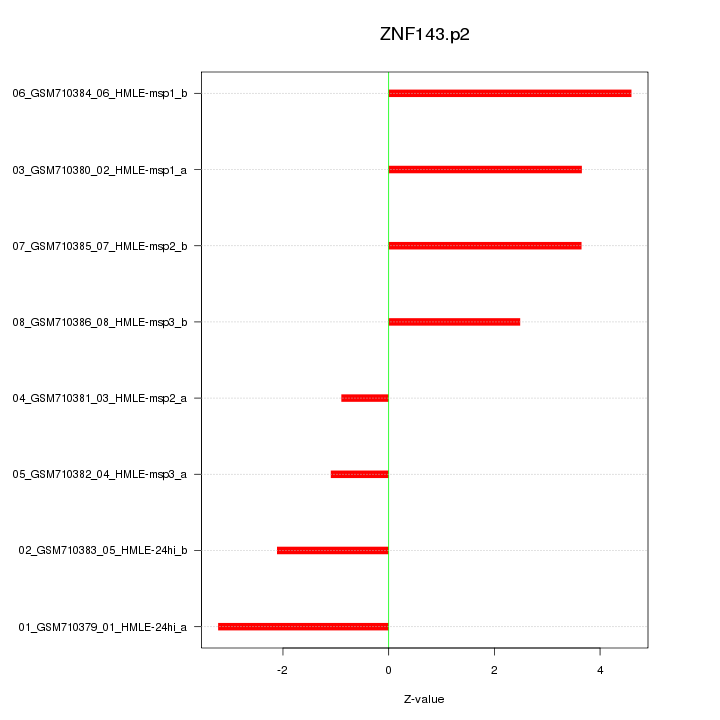
|
chr2_+_202316391
|
1.828
|
NM_001206864
NM_018571
|
STRADB
|
STE20-related kinase adaptor beta
|
|
chr19_-_44123732
|
1.817
|
|
ZNF428
|
zinc finger protein 428
|
|
chr22_-_30162933
|
1.552
|
NM_001003692
NM_019103
|
ZMAT5
|
zinc finger, matrin-type 5
|
|
chr19_-_37958338
|
1.374
|
NM_152484
|
ZNF569
|
zinc finger protein 569
|
|
chr15_-_41099645
|
1.290
|
|
DNAJC17
|
DnaJ (Hsp40) homolog, subfamily C, member 17
|
|
chr2_-_202316227
|
1.276
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr15_-_41099669
|
1.211
|
NM_018163
|
DNAJC17
|
DnaJ (Hsp40) homolog, subfamily C, member 17
|
|
chr19_-_48673559
|
1.181
|
NM_000234
|
LIG1
|
ligase I, DNA, ATP-dependent
|
|
chr14_+_104394751
|
1.166
|
NM_153046
|
TDRD9
|
tudor domain containing 9
|
|
chr18_-_77439267
|
1.040
|
|
|
|
|
chr12_-_88535746
|
1.038
|
NM_025114
|
CEP290
|
centrosomal protein 290kDa
|
|
chr19_+_14544100
|
1.032
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr11_-_118550354
|
1.023
|
NM_007180
|
TREH
|
trehalase (brush-border membrane glycoprotein)
|
|
chr19_+_14551080
|
1.018
|
NM_213560
|
PKN1
|
protein kinase N1
|
|
chr19_+_14551052
|
1.007
|
|
PKN1
|
protein kinase N1
|
|
chr16_+_2802634
|
1.002
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr8_-_38326351
|
0.993
|
NM_001174063
NM_001174064
NM_015850
NM_023105
NM_023106
NM_023110
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr19_-_58459013
|
0.992
|
NM_005773
|
ZNF256
|
zinc finger protein 256
|
|
chr10_+_38383218
|
0.978
|
NM_001007094
NM_001178101
NM_003421
|
ZNF37A
|
zinc finger protein 37A
|
|
chr19_+_14551087
|
0.976
|
|
PKN1
|
protein kinase N1
|
|
chr20_+_62694456
|
0.954
|
|
TCEA2
|
transcription elongation factor A (SII), 2
|
|
chr5_-_171433605
|
0.954
|
|
FBXW11
|
F-box and WD repeat domain containing 11
|
|
chr8_+_17354562
|
0.943
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr17_+_76210329
|
0.932
|
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr17_+_76210276
|
0.913
|
NM_001012270
NM_001012271
NM_001168
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr19_+_54024176
|
0.904
|
NM_018555
|
ZNF331
|
zinc finger protein 331
|
|
chr2_+_201450730
|
0.883
|
NM_001159
|
AOX1
|
aldehyde oxidase 1
|
|
chr8_-_38326115
|
0.882
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr19_+_15218141
|
0.864
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr11_-_134123138
|
0.852
|
NM_001037304
NM_001037305
NM_014174
NM_199297
NM_199298
|
THYN1
|
thymocyte nuclear protein 1
|
|
chr1_+_156737315
|
0.847
|
|
PRCC
|
papillary renal cell carcinoma (translocation-associated)
|
|
chr10_-_79397394
|
0.841
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chrX_-_106960268
|
0.834
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr10_-_44070008
|
0.824
|
NM_001099282
NM_001099283
NM_001099284
|
ZNF239
|
zinc finger protein 239
|
|
chr19_+_507475
|
0.823
|
NM_033513
|
TPGS1
|
tubulin polyglutamylase complex subunit 1
|
|
chr19_+_4304590
|
0.808
|
NM_024333
|
FSD1
|
fibronectin type III and SPRY domain containing 1
|
|
chr11_-_65488142
|
0.799
|
NM_032193
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr11_+_134123406
|
0.798
|
NM_014384
|
ACAD8
|
acyl-CoA dehydrogenase family, member 8
|
|
chr19_-_44124013
|
0.796
|
NM_182498
|
ZNF428
|
zinc finger protein 428
|
|
chr1_+_156737273
|
0.777
|
NM_005973
|
PRCC
|
papillary renal cell carcinoma (translocation-associated)
|
|
chr20_+_46130656
|
0.772
|
|
NCOA3
|
nuclear receptor coactivator 3
|
|
chrX_-_101771641
|
0.755
|
NM_021992
|
TMSB15A
TMSB15B
|
thymosin beta 15a
thymosin beta 15B
|
|
chr17_-_76921453
|
0.753
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr1_+_156024516
|
0.753
|
NM_001145264
NM_014017
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2
|
|
chr19_+_12917391
|
0.750
|
|
RNASEH2A
|
ribonuclease H2, subunit A
|
|
chr19_-_2456954
|
0.747
|
NM_032737
|
LMNB2
|
lamin B2
|
|
chr5_-_171433876
|
0.744
|
NM_012300
NM_033644
NM_033645
|
FBXW11
|
F-box and WD repeat domain containing 11
|
|
chr4_-_8160435
|
0.737
|
NM_001130083
NM_001130084
NM_001130085
NM_001130086
NM_001130087
NM_001130088
NM_032432
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr12_-_29936685
|
0.734
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr1_-_85930733
|
0.733
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr1_+_40974430
|
0.732
|
NM_022774
|
DEM1
|
defects in morphology 1 homolog (S. cerevisiae)
|
|
chr2_-_242447449
|
0.730
|
|
STK25
|
serine/threonine kinase 25
|
|
chr4_+_1723212
|
0.728
|
NM_006342
|
TACC3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr16_-_31106049
|
0.724
|
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr1_-_1293907
|
0.712
|
NM_032348
|
MXRA8
|
matrix-remodelling associated 8
|
|
chr5_-_171433502
|
0.711
|
|
FBXW11
|
F-box and WD repeat domain containing 11
|
|
chr1_-_186344383
|
0.705
|
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chr14_-_77923888
|
0.703
|
NM_001193314
NM_001193315
NM_001193316
NM_001193317
NM_022067
|
C14orf133
|
chromosome 14 open reading frame 133
|
|
chr16_+_2802683
|
0.698
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr19_+_44556163
|
0.689
|
NM_013361
|
ZNF223
|
zinc finger protein 223
|
|
chr11_-_73309090
|
0.688
|
NM_015159
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr22_-_41682169
|
0.687
|
|
RANGAP1
|
Ran GTPase activating protein 1
|
|
chr7_+_44144002
|
0.683
|
|
AEBP1
|
AE binding protein 1
|
|
chr16_+_3074027
|
0.681
|
NM_001142350
NM_024339
|
THOC6
|
THO complex 6 homolog (Drosophila)
|
|
chr16_-_3184858
|
0.679
|
|
|
|
|
chr22_-_41682187
|
0.679
|
NM_002883
|
RANGAP1
|
Ran GTPase activating protein 1
|
|
chrX_-_48931645
|
0.678
|
NM_007213
|
PRAF2
|
PRA1 domain family, member 2
|
|
chr20_+_46130600
|
0.676
|
NM_001174087
NM_001174088
NM_006534
NM_181659
|
NCOA3
|
nuclear receptor coactivator 3
|
|
chr16_-_31106125
|
0.660
|
NM_024006
NM_206824
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr16_+_3074041
|
0.658
|
|
THOC6
|
THO complex 6 homolog (Drosophila)
|
|
chr12_+_107168336
|
0.658
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr16_-_31106093
|
0.648
|
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr6_-_30710219
|
0.647
|
NM_005803
|
FLOT1
|
flotillin 1
|
|
chr20_+_30945988
|
0.647
|
NM_001164603
NM_015338
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr4_-_185747051
|
0.640
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr10_+_104613966
|
0.640
|
NM_001136200
NM_144591
|
C10orf32-AS3MT
C10orf32
|
C10orf32-AS3MT readthrough
chromosome 10 open reading frame 32
|
|
chr5_+_178450775
|
0.636
|
NM_001136116
|
ZNF879
|
zinc finger protein 879
|
|
chr20_+_32399046
|
0.634
|
NM_176812
|
CHMP4B
|
charged multivesicular body protein 4B
|
|
chr10_-_38265452
|
0.632
|
NM_145011
|
ZNF25
|
zinc finger protein 25
|
|
chr22_+_44319618
|
0.628
|
NM_025225
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr8_+_72756218
|
0.627
|
|
LOC100132891
|
uncharacterized LOC100132891
|
|
chr16_+_3074051
|
0.624
|
|
THOC6
|
THO complex 6 homolog (Drosophila)
|
|
chr6_-_27440459
|
0.623
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr19_-_38270199
|
0.617
|
NM_001172689
NM_001172690
NM_001172691
NM_001172692
NM_152360
|
ZNF573
|
zinc finger protein 573
|
|
chr16_+_19535178
|
0.609
|
NM_001199022
NM_014711
|
CCP110
|
centriolar coiled coil protein 110kDa
|
|
chr19_-_1401474
|
0.607
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr16_+_3074060
|
0.604
|
|
THOC6
|
THO complex 6 homolog (Drosophila)
|
|
chr5_-_114505549
|
0.602
|
|
TRIM36
|
tripartite motif containing 36
|
|
chr12_-_124457099
|
0.601
|
NM_025140
|
CCDC92
|
coiled-coil domain containing 92
|
|
chr8_+_72756335
|
0.599
|
|
|
|
|
chr16_+_2802605
|
0.598
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chrX_-_106960028
|
0.595
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr19_+_58816714
|
0.594
|
|
ERVK3-1
|
endogenous retrovirus group K3, member 1
|
|
chr16_+_2802656
|
0.590
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr20_-_48732479
|
0.587
|
NM_022442
NM_021988
NM_199144
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr10_-_15210610
|
0.586
|
NM_004808
|
NMT2
|
N-myristoyltransferase 2
|
|
chr4_-_185747183
|
0.583
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr20_+_32399128
|
0.582
|
|
CHMP4B
|
charged multivesicular body protein 4B
|
|
chr22_+_44319674
|
0.580
|
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr1_+_244816379
|
0.580
|
|
PPPDE1
|
PPPDE peptidase domain containing 1
|
|
chr7_-_137531585
|
0.576
|
NM_004717
|
DGKI
|
diacylglycerol kinase, iota
|
|
chrX_-_106959597
|
0.575
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr6_-_111136330
|
0.572
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr19_+_58816757
|
0.568
|
|
ERVK3-1
|
endogenous retrovirus group K3, member 1
|
|
chr2_+_148778573
|
0.566
|
NM_018328
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr19_+_12917409
|
0.566
|
NM_006397
|
RNASEH2A
|
ribonuclease H2, subunit A
|
|
chr15_+_63481713
|
0.565
|
NM_016530
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr17_-_38083762
|
0.558
|
NM_139280
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae)
|
|
chr19_+_44617510
|
0.556
|
NM_013362
|
ZNF225
|
zinc finger protein 225
|
|
chr19_-_44439410
|
0.551
|
NM_003425
|
ZNF45
|
zinc finger protein 45
|
|
chr16_+_2802673
|
0.550
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr8_+_16884745
|
0.550
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr12_+_133562941
|
0.549
|
NM_019591
|
ZNF26
|
zinc finger protein 26
|
|
chr11_+_58874614
|
0.547
|
NM_001142703
NM_001142704
NM_198947
|
FAM111B
|
family with sequence similarity 111, member B
|
|
chr19_+_57050299
|
0.543
|
NM_020828
|
ZFP28
|
zinc finger protein 28 homolog (mouse)
|
|
chr20_-_18477811
|
0.540
|
NM_006606
|
RBBP9
|
retinoblastoma binding protein 9
|
|
chr11_+_62104882
|
0.539
|
|
ASRGL1
|
asparaginase like 1
|
|
chr1_-_26324534
|
0.536
|
NM_000437
|
PAFAH2
|
platelet-activating factor acetylhydrolase 2, 40kDa
|
|
chr2_+_25016174
|
0.533
|
NM_001199803
NM_024322
|
CENPO
|
centromere protein O
|
|
chr6_+_56911422
|
0.533
|
|
KIAA1586
|
KIAA1586
|
|
chr1_-_1551041
|
0.528
|
|
MIB2
|
mindbomb homolog 2 (Drosophila)
|
|
chr14_+_24025188
|
0.525
|
NM_001126339
NM_024328
|
THTPA
|
thiamine triphosphatase
|
|
chr3_+_49449638
|
0.525
|
NM_022171
|
TCTA
|
T-cell leukemia translocation altered gene
|
|
chr13_-_67804432
|
0.523
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr9_-_99540327
|
0.520
|
NM_014930
|
ZNF510
|
zinc finger protein 510
|
|
chr12_+_133656984
|
0.514
|
NM_003440
|
ZNF140
|
zinc finger protein 140
|
|
chr8_+_27631902
|
0.511
|
NM_001017420
|
ESCO2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae)
|
|
chr22_+_30163351
|
0.510
|
NM_001003684
NM_013387
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr3_+_154798078
|
0.510
|
NM_007289
|
MME
|
membrane metallo-endopeptidase
|
|
chr14_+_65453605
|
0.507
|
|
FNTB
|
farnesyltransferase, CAAX box, beta
|
|
chr3_+_51976274
|
0.506
|
NM_001003931
NM_005485
|
PARP3
|
poly (ADP-ribose) polymerase family, member 3
|
|
chr12_+_12764844
|
0.505
|
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr15_-_78369825
|
0.505
|
NM_015079
NM_144572
|
TBC1D2B
|
TBC1 domain family, member 2B
|
|
chr4_-_159592979
|
0.502
|
NM_001008393
|
C4orf46
|
chromosome 4 open reading frame 46
|
|
chr2_-_27593312
|
0.499
|
NM_001034116
NM_015636
|
EIF2B4
|
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa
|
|
chr16_-_686292
|
0.498
|
NM_001040160
NM_001040161
NM_001040162
NM_001040165
NM_032366
|
C16orf13
|
chromosome 16 open reading frame 13
|
|
chr13_-_41768533
|
0.497
|
NM_032138
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr11_-_62314291
|
0.496
|
NM_001620
NM_024060
|
AHNAK
|
AHNAK nucleoprotein
|
|
chr14_+_55034633
|
0.496
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr15_-_91475653
|
0.496
|
NM_198527
|
HDDC3
|
HD domain containing 3
|
|
chr3_-_49466663
|
0.493
|
|
NICN1
|
nicolin 1
|
|
chr19_+_35168558
|
0.493
|
NM_001012320
NM_018443
|
ZNF302
|
zinc finger protein 302
|
|
chr1_+_203764802
|
0.491
|
|
ZC3H11A
|
zinc finger CCCH-type containing 11A
|
|
chr19_+_37096202
|
0.488
|
NM_032825
|
ZNF382
|
zinc finger protein 382
|
|
chr7_-_111846384
|
0.487
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr17_+_38296506
|
0.486
|
NM_007359
|
CASC3
|
cancer susceptibility candidate 3
|
|
chr1_+_159175048
|
0.486
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr6_-_111136407
|
0.484
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr19_-_12845539
|
0.484
|
|
C19orf43
|
chromosome 19 open reading frame 43
|
|
chr10_-_31320772
|
0.481
|
NM_001143766
NM_001143767
NM_001143768
NM_001143770
NM_001143771
NM_182755
|
ZNF438
|
zinc finger protein 438
|
|
chr17_-_1619473
|
0.477
|
|
MIR22HG
|
MIR22 host gene (non-protein coding)
|
|
chr3_-_179169180
|
0.475
|
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr10_+_95848811
|
0.474
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr19_+_35225095
|
0.473
|
|
ZNF181
|
zinc finger protein 181
|
|
chr4_+_1723233
|
0.472
|
|
TACC3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr22_+_44319685
|
0.472
|
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr12_-_102455739
|
0.471
|
|
CCDC53
|
coiled-coil domain containing 53
|
|
chr9_-_40792062
|
0.466
|
NM_033160
|
ZNF658
|
zinc finger protein 658
|
|
chr7_+_44143945
|
0.464
|
NM_001129
|
AEBP1
|
AE binding protein 1
|
|
chr16_+_2802676
|
0.463
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr22_+_21336616
|
0.461
|
|
LZTR1
|
leucine-zipper-like transcription regulator 1
|
|
chr6_-_105307747
|
0.459
|
|
HACE1
|
HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1
|
|
chr19_-_38714781
|
0.459
|
NM_001135155
NM_004647
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr19_+_925984
|
0.457
|
NM_005224
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like)
|
|
chr19_-_54619025
|
0.454
|
NM_013342
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia)
|
|
chr4_+_145567147
|
0.450
|
NM_022475
|
HHIP
|
hedgehog interacting protein
|
|
chr7_+_7606634
|
0.448
|
|
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila)
|
|
chr4_+_123747815
|
0.448
|
NM_002006
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr20_-_47804903
|
0.448
|
NM_001037328
NM_004602
NM_017452
NM_017453
NM_017454
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chr19_+_37862055
|
0.448
|
NM_032453
|
ZNF527
|
zinc finger protein 527
|
|
chr17_-_76921205
|
0.447
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr19_+_37096225
|
0.446
|
|
ZNF382
|
zinc finger protein 382
|
|
chr19_-_37406918
|
0.446
|
NM_001171979
|
ZNF829
|
zinc finger protein 829
|
|
chr21_-_47648685
|
0.443
|
|
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase)
|
|
chr22_-_36903033
|
0.443
|
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr11_-_74109417
|
0.441
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr15_+_63481788
|
0.441
|
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr7_+_7222142
|
0.440
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr20_-_47804675
|
0.438
|
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chr6_-_111136616
|
0.437
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr19_+_56915382
|
0.435
|
NM_001159860
|
ZNF583
|
zinc finger protein 583
|
|
chr19_+_37341259
|
0.434
|
NM_003419
|
ZNF345
|
zinc finger protein 345
|
|
chr1_-_243418675
|
0.433
|
NM_001042404
NM_001042405
NM_014812
|
CEP170
|
centrosomal protein 170kDa
|
|
chr16_-_30007315
|
0.432
|
NM_001197323
NM_003609
|
HIRIP3
|
HIRA interacting protein 3
|
|
chr12_-_46384317
|
0.431
|
|
SCAF11
|
SR-related CTD-associated factor 11
|
|
chr19_-_44617271
|
0.431
|
|
LOC100379224
|
uncharacterized LOC100379224
|
|
chr11_-_82782883
|
0.431
|
NM_014488
|
RAB30
|
RAB30, member RAS oncogene family
|
|
chr1_-_26232643
|
0.430
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr20_+_62289640
|
0.429
|
|
RTEL1
|
regulator of telomere elongation helicase 1
|
|
chr15_-_40987284
|
0.426
|
|
LOC100505648
|
uncharacterized LOC100505648
|
|
chr9_-_116037683
|
0.425
|
|
FKBP15
|
FK506 binding protein 15, 133kDa
|
|
chr19_-_36909463
|
0.424
|
NM_133466
|
ZFP82
|
zinc finger protein 82 homolog (mouse)
|
|
chr1_-_26232890
|
0.424
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr16_+_67207755
|
0.422
|
NM_003946
NM_001185057
|
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain)
|
|
chr14_+_65453439
|
0.422
|
NM_002028
|
FNTB
|
farnesyltransferase, CAAX box, beta
|
|
chr5_+_159436155
|
0.421
|
NM_003314
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr7_-_150754979
|
0.420
|
NM_001164410
NM_004935
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr7_-_150754934
|
0.419
|
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr16_+_69599869
|
0.417
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr11_-_35547144
|
0.415
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|